Published online Apr 14, 2014. doi: 10.3748/wjg.v20.i14.3858
Revised: January 9, 2014
Accepted: January 19, 2014
Published online: April 14, 2014
Processing time: 197 Days and 16.2 Hours
Colorectal cancer (CRC) is one of the most common human malignant diseases and the second leading cause of cancer-related deaths worldwide. The treatment of advanced CRC has improved significantly in recent years. With the emergence of two targeted antibodies, cetuximab (Erbitux), an anti-epidermal growth factor receptor monoclonal antibody and bevacizumab (Avastin), a vascular endothelial growth factor monoclonal antibody, the treatment of metastatic CRC has entered the era of personalized therapy. Predictive and prognostic biomarkers have, and will continue to, facilitate the selection of suitable patients and the personalization of treatment for metastatic CRC (mCRC). In this review, we will focus primarily on the important progresses made in the personalized treatment of mCRC and discuss the potentially novel predictive and prognostic biomarkers for improved selection of patients for anti-cancer treatment in the future.
Core tip: This review focuses primarily on the important progresses achieved in the personalized treatment of metastatic colorectal cancer and highlights the potentially novel predictive and prognostic biomarkers for improved selection of patients for anti-cancer treatment in the future.
- Citation: Luo HY, Xu RH. Predictive and prognostic biomarkers with therapeutic targets in advanced colorectal cancer. World J Gastroenterol 2014; 20(14): 3858-3874
- URL: https://www.wjgnet.com/1007-9327/full/v20/i14/3858.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i14.3858
Colorectal cancer (CRC) is one of the most common human malignancies and the second leading cause of cancer-related deaths worldwide. There are 25159 new cases of CRC diagnosed each year and 12161 CRC-related deaths in China[1]. Metastasis to the liver and lung are the main cause of death, with approximately 40%-50% of all patients experiencing metastasis[2,3]. The treatment of advanced CRC has improved significantly in recent years, and the overall survival (OS) for metastatic CRC (mCRC) patients has increased from a median of 10 mo to more than 20 mo[4]. With the emergence of two targeted antibodies, cetuximab (Erbitux), an anti-epidermal growth factor receptor (EGFR) monoclonal antibody and bevacizumab (Avastin), a vascular endothelial growth factor (VEGF) monoclonal antibody, the treatment of mCRC has entered the era of personalized therapy. Treatment on a “personalized” basis now involves a simultaneous case-specific analysis of clinical and pathological characteristics and analysis of a patient’s genetic and tumor biomarker profile. Predictive and prognostic biomarkers have, and will continue to, facilitate the selection of suitable patients and the personalization of treatment for mCRC.
A prognostic factor is defined as any parameter, evaluated at diagnosis (or surgery), which is associated with treatment outcome (disease-free interval, survival, local control) and may predict patient outcome independent of treatment. Prognostic factors (biological or clinical) may be defined at any disease stage or setting (for example, performance status in the advanced disease setting). A predictive factor is any parameter which identifies patients who will benefit from a particular treatment and evaluates the response or lack of response to specific treatment. Over the last 30 years, there has been significant advancement in understanding the molecular origins of CRC and the characteristics of tumor aggressiveness[5]. However, in practice, the distinction between prognostic and predictive factors is not straightforward, and many factors are a mixture of the two. Understanding the molecular mechanisms underlying the metastatic process will help us to identify those at the highest risk of recurrence and to find new tumor targets to prevent disease progression.
This review focuses primarily on the important progresses made in the personalized treatment of mCRC and highlights the potentially novel predictive and prognostic biomarkers for improved selection of patients for the anti-cancer treatment in the future.
The appropriate use of targeted biologic agents can positively impact a patient’s prognosis. Extensive research has focused on tumor factors due to the central role they play in the response to targeted biologic agents. Currently, numerous potential biomarkers are under investigation, and these biomarkers may be clinically useful in the future once validated by appropriate trials (Table 1).
| Biomarker | Prevalence | Evidence available | Predictive and prognostic value |
| KRAS mutations | 40% | Conclusive | Negative predictive biomarker for anti-EGFR mAbs |
| Insufficient | Predicts poor prognosis, but not an independent prognostic factor | ||
| BRAF mutations | 10% | Substantial | Prognostic marker for poor outcome |
| Insufficient | Potential predictive marker for resistance to anti-EGFR mAbs | ||
| NRAS mutations | 3%-5% | Insufficient1 | Potential predictive marker for resistance to anti-EGFR mAbs |
| PIK3CA mutations | 15%-20% | Insufficient1 | Potential predictive marker for resistance to cetuximab (exon 20, not exon 9 mutations) |
| Potential prognostic marker for poor outcome | |||
| PTEN (loss of expression) | 20%-40% | Insufficient1 | Potential predictive marker for resistance to cetuximab |
| Associated with activation of the PIK3CA pathway and adverse disease outcome | |||
| P53 mutations | 1%-5% | Insufficient1 | An independent predictive factor for cetuximab benefit |
| Not prognostic | |||
| Epiregulin, amphiregulin (high expression) | 50%-60% | Insufficient1 | Associated with resistance to anti-EGFR antibody therapy and adverse clinical outcome |
| VEGF-D | 40%-75% | Insufficient1 | Potential predictive marker for response to bevacizumab |
| VEGF-A | Insufficient1 | Not predictive of response to bevacizumab |
An important molecular target for mCRC treatment is the epidermal growth factor receptor (EGFR). EGFR is a receptor tyrosine kinase frequently expressed in epithelial tumors. Binding of a ligand to the extracellular domain of EGFR activates intracellular signalling via several pathways, including the RAS/RAF/MAPK pathway and the PI3K/Akt axis[6]. EGFR is expressed on normal human cells, but higher levels of expression have also been correlated with malignancy in a variety of cancers, including CRC[7]. EGFR has been implicated in colorectal tumorigenesis, tumor progression, and metastasis[8,9]. EGFR is overexpressed in 30%-85% of patients with CRC and has been associated with advanced stage disease. Numerous studies have evaluated the prognostic relevance of EGFR in CRC, but the impact of its expression on survival remains controversial[10]. Two monoclonal antibodies, cetuximab (Erbitux™; Bristol Myers Squibb, Inc., Princeton, NJ, United States) and panitumumab (Vectibix™; Amgen, Inc., Thousand Oaks, CA, United States), target the human EGFR in the treatment of EGFR-overexpressing CRC[11,12]. Genetic alterations of EGFR and its downstream signaling effectors may predict response to anti-EGFR monoclonal antibodies (mAbs), therefore research efforts have been made to understand the specific resistance mechanisms.
The main research areas in this setting have focused on the role of (1) EGFR protein expression; (2) EGFR gene copy number; (3) EGFR gene mutations; (4) overexpression of EGFR ligands (such as epiregulin and amphiregulin); and (5) markers of EGFR downstream signaling[13-17].
Overexpression of EGFR protein, as determined by immunohistochemistry (IHC), was initially selected as an entry criterion for early studies evaluating EGFR inhibitors on the assumption that sensitivity to such agents was associated with EGFR expression[18]. However, a large body of evidence from mCRC patients who were treated with anti-EGFR mAbs[19-21] indicates that this biomarker is poorly associated with response. Moreover, several authors reported that cetuximab was also active in EGFR-negative tumors detected by IHC[22,23]. EGFR expression at either the protein or mRNA level is not correlated with anti-EGFR mAbs response.
In a small fraction of CRCs, EGFR overexpression is frequently associated with amplification of the gene (17% in primary and 23% in metastatic tumors)[24]. Activating mutations in the EGFR catalytic domain are seen frequently in lung cancer and play an important role in determining responsiveness to anti-EGFR therapy[25]. However, EGFR mutations are very rare in CRC and are not significantly associated with response to anti-EGFR mAbs treatment[26,27].
In contrast, increased EGFR gene copy number (EGFR GCN) has been associated with response to anti-EGFR therapy and with prognosis of mCRC in small retrospective studies[28,29]. Recently, Yang et al[30] performed a meta-analysis to summarize the evidence for the predictive value of EGFR GCNfor clinical outcomes of mCRC patients treated with anti-EGFR mAbs. The data showed that increased EGFR GCNwas generally associated with a better objective response, especially among patients with wild-type KRAS. In another meta-analysis performed by Jiang et al[31], increased EGFR GCN was significantly associated with improved OS and progression-free survival (PFS) in the population that received second-line or higher therapy. The prognostic impact of EGFR GCN on survival does not appear to be related to KRAS status, which suggests that EGFR GCN might be an independent prognostic biomarker. EGFR GCN can be detected by fluorescence in situ hybridization (FISH), chromogenic in situ hybridization (CISH) or polymerase chain reaction (PCR)-based methods. Interestingly, the EGFR GCN evaluated by quantitative PCR does not appear to correlate with the clinical outcome of patients, whereas the results of FISH analysis appear to be associated with an increase in treatment response[32]. The comparability of these methods and their differential impact on results still needs to be defined. However, EGFR copy number is not used in clinical practice to select patients for treatment, partly due to the lack of standardization of FISH technology and the uncertainty of published clinical cutoff values. Further studies are required to assess the increase in EGFR GCN as a predictive biomarker of response to anti-EGFR therapy.
Increased expression of alternative EGFR ligands, such as amphiregulin and epiregulin, may promote tumor growth via an autocrine or paracrine loop that signals through EGFR and have been shown in retrospective studies to be predictive of response to cetuximab[33-35]. The level of sensitivity to cetuximab was shown to be proportional to the intensity of epiregulin and amphiregulin mRNA expression[35-38]. Two studies demonstrated that mCRC patients with KRAS wild-type tumors and high amphiregulin and epiregulin mRNA expression were more likely to have disease control with cetuximab treatment[35,37]. In addition to their predictive value, amphiregulin and epiregulin mRNA expression appears to be a useful prognostic marker in KRAS wild-type patients regardless of whether they were receiving anti-EGFR therapy[39]. Low expression of EGFR activating ligands, amphiregulin and epiregulin, was associated with resistance to anti-EGFR therapy and adverse clinical outcome, however, these ligands are not routinely measured in clinical practice and further evaluation of their role is required.
In brief, the predictive value of EGFR expression remains unconvincing in the use of anti-EGFR therapy. Therefore, the focus has shifted to alterations in the key signaling pathway downstream of EGFR, which may drive the growth and progression of CRC and provide an escape mechanism that allows tumors to overcome the pharmacological blockade induced by anti-EGFR mAbs. KRAS, BRAF, PTEN, and PI3KCA mutations have been highlighted as the mechanisms that activate the EGFR signaling pathway.
KRAS belongs to the rat sarcoma virus (ras) gene family of oncogenes which includes KRAS, HRAS, and NRAS. All of these oncogenes when mutated have the ability to transform cells, but KRAS is the most commonly mutated RAS family member in CRC[40]. KRAS mutations occur in approximately 35%-45% of mCRC patients, and lead to the constitutive activation of EGFR downstream pathways[3]. KRAS mutation is thought to be a fairly early event in colon carcinogenesis and appears to be ≥ 95% concordant between primary tumor and metastatic sites[41-43]. Point mutations in KRAS occur most frequently in codons 12, 13 (exon 2), 61 (exon 3)[44], and 146 (exon 4)[45], and up to 90% of activating KRAS gene mutations are detected in codons 12 (82%-87%) and 13 (13%-18%). These are generally observed as somatic mutations.
A number of studies have evaluated the potential prognostic role of KRAS in CRCs, but the data are conflicting largely due to the differences in methodology and datasets analyzed[46-50]. The first RASCAL meta-analysis evaluated the KRAS gene status in 2721 patients, and suggested that the presence of a mutation increased the risk of recurrence (P < 0.001) and death (P = 0.004)[46]. This finding was later restricted to the G12V mutation, which had a statistically significant impact on treatment failure-free survival (HR = 1.3, P = 0.004) and OS (HR = 1.29, P = 0.008)[47]. Furthermore, the N0147 trial which evaluated the treatment with cetuximab combined with FOLFOX in patients with resected stage III CRC showed that the 3-year disease-free survival in patients with wild-type KRAS was significantly better than that in patients with KRAS mutations (72.3% vs 64.2%, HR = 0.7, P = 0.004). These analyses suggest that KRAS mutations are independent prognostic factors[51]. The COIN trial assessed the effects of cetuximab combined with oxaliplatin and fluoropyrimidine chemotherapy as first-line treatment in patients with advanced CRC. This trial also showed that a KRAS mutation was a strong negative prognostic factor, and the median OS was significantly shorter in patients with KRAS, NRAS, or BRAF mutations (n = 706, 13.6 mo) compared to those with wild-type KRAS, NRAS, and BRAF (n = 581, 20.1 mo), irrespective of treatment[52].
However, a recent study by Roth et al[53] suggested that the prognostic value of KRAS mutation status for PFS and OS was lacking in large adjuvant trials of patients with stage II and III resected colon cancer. Investigators from the PETACC-3 trial retrospectively analyzed archival tissue (n = 1564) for mutations in KRAS (exon 2, codons 12 and 13) and found no clear association with relapse-free survival (RFS) or OS in both univariate and multivariate analyses. In the CALGB 89803 study[54], stage III CRC patients with KRAS mutated tumors did not experience any difference in DFS, RFS and OS rates compared to patients with KRAS wild-type tumors.
In advanced CRC, a few phase 3 studies comparing cetuximab[55] or panitumumab[20,56] with best supportive care (BSC) in the third-line setting demonstrated no significant prognostic value based on KRAS mutation status. Two large studies evaluating the addition of cetuximab or panitumumab to chemotherapy and bevacizumab in the first-line setting did not find a prognostic value for KRAS mutational status[57,58].
It may be difficult to interpret the various studies published on the prognostic role of KRAS. Therefore, further prospective studies are required to confirm whether a specific KRAS mutation might lead to a clinically relevant prognostic effect in patients with CRC.
The predictive value of KRAS has been investigated extensively in the era of EGFR-targeted therapy in colon cancer. Evidence from several clinical trials demonstrated that KRAS mutations have emerged as a major predictor of resistance to anti-EGFR mAbs in CRC. Several retrospective analyses have been conducted to explore the role of KRAS mutations as a negative predictive biomarker of tumors in patients with mCRC treated with anti-EGFR antibody (with or without chemotherapy)[13,55,59]. The first study to evaluate the correlation between K-RAS mutational status and lack of response to treatment with cetuximab was performed by Lièvre et al[59]. They analyzed 30 patients predominantly treated with cetuximab plus irinotecan after previous exposure to chemotherapy, and KRAS mutations were observed in 13 of the 30 patients enrolled (43%). None of the mutated tumors responded to cetuximab treatment. The OS of KRAS wild-type patients was significantly higher compared to those with mutated KRAS. The negative predictive value of KRAS mutations for response to anti-EGFR therapy has been confirmed in a number of single arm retrospective studies using the EGFR inhibitors cetuximab or panitumumab alone or in combination with chemotherapy. These retrospective studies revealed that patients with KRAS mutations receiving first and subsequent lines of treatment do not benefit from anti-EGFR therapy, and that they show no survival benefit from such treatments[13,59].
Data from phase III trials using anti-EGFR targeted therapy in the metastatic setting also suggested that mutated KRAS status predicts a lack of response[20,60,61]. The biomarker analysis of the pivotal phase III trial of panitumumab monotherapy in the relapsed or refractory setting was the first large study (n = 463 patients) to confirm the negative predictive value of KRAS mutations[20]. This study found that in those treated with panitumumab, PFS was 12.3 wk in the subgroup of patients with the wild-type KRAS gene, but only 7.4 wk in the subgroup of patients with the mutant KRAS gene. This was statistically significant. The PRIME trial evaluated the addition of panitumumab to FOLFOX4 for the initial treatment of patients with KRAS wild-type mCRC[62]. The results were prospectively analyzed by tumor KRAS status, which demonstrated a significantly longer PFS when panitumumab was added to chemotherapy in patients with KRAS wild-type tumors (9.6 mo vs 8 mo, respectively; HR = 0.80, 95%CI: 0.66-0.97, P = 0.02). Furthermore, additional phase III studies have shown that only patients with KRAS wild-type CRC will benefit from the addition of panitumumab to FOLFIRI as second-line treatment[63].
Data have recently been published from two large randomized phase II-III studies carried out to examine the benefits of cetuximab as first-line treatment for mCRC[60,61]. The CRYSTAL study demonstrated that only patients with wild-type KRAS tumors benefited from the addition of cetuximab to FOLFIRI, showing a higher response rate (RR) (57.3% vs 39.7%, P < 0.0001), longer PFS (median, 9.9 mo vs 8.4 mo, P = 0.012) and longer OS (median, 23.5 mo vs 20.0 mo, P = 0.0093). In patients whose tumors carried KRAS mutations, there was no evidence of benefit associated with the addition of cetuximab to FOLFIRI. The OPUS trial also showed that the addition of cetuximab to the FOLFOX-4 regimen was only beneficial in the wild-type KRAS subgroup[61]. In KRAS wild-type patients, the addition of cetuximab to FOLFOX induced a significant increase in RR (61% vs 37%; P = 0.011) and PFS (7.7 mo vs 7.2 mo, HR = 0.57, P = 0.0163) without OS benefit. In contrast, a negative impact on treatment efficacy was noted when cetuximab was added to chemotherapy in patients with KRAS mutant mCRC[64]. These results indicate that KRAS mutated patients do not benefit from the addition of cetuximab to conventional chemotherapy.
In contrast to these results, other phase III trials found that KRAS mutation status was not predictive of benefit when cetuximab was combined with first-line chemotherapy[52,65]. In the NORDIC VII trial, cetuximab combined with the continuous or intermittent FLOX regimen [bolus 5-fluorouracil (5-FU) plus oxaliplatin] did not significantly improve efficacy compared with FLOX alone[65]. In the large COIN trial, the addition of cetuximab to oxaliplatin-based chemotherapy did not benefit OS or PFS in KRAS wild-type patients[52].
When anti-EGFR therapy was added to bevacizumab-based first-line chemotherapy in advanced CRC, no additional benefit was observed, even in patients with wild-type KRAS tumors[57,58]. In the CAIRO-2 study, the addition of cetuximab to capecitabine, oxaliplatin, and bevacizumab as first-line treatment in patients with mCRC had no effect on RR (50% vs 61.4%; P = 0.06) or PFS (median, 10.5 vs 10.6; P = 0.3) among those with tumors carrying wild-type KRAS. Similarly, in the PACCE study, the addition of panitumumab to bevacizumab and oxaliplatin-based chemotherapy was associated with shorter PFS and OS in patients with tumors carrying wild-type KRAS. These data suggest a detrimental effect following the addition of antiangiogenic agents to anti-EGFR therapies in advanced CRC.
Based on current information from these clinical trials, the guidelines of the National Comprehensive Cancer Network (NCCN), the ESMO (European Society for Medical Oncology), and the ASCO recommend the use of anti-EGFR-directed therapy only in mCRC patients with wild-type KRAS status. In addition, the NCCN guideline also recommends testing for KRAS mutations in codons 12 and 13 in certified laboratories. This is the first true use of personalized medicine in CRC.
However, it is interesting that not all KRAS mutations are equal in their biological characteristics and their impact on mediating EGFR resistance. Anecdotal reports indicate that a very small number of patients (< 10%) with KRAS-mutated tumors respond to anti-EGFR therapy[13,66,67] and that about 15% have long-term disease stabilization[68]. Preclinical data demonstrated that cell lines with KRAS codon 13 glycine-to-aspartate (G13D) mutations exhibit weaker in vitro transforming activity than codon 12 mutations[69,70]. Moreover, a recently published retrospective pooled exploratory analysis of patients with chemotherapy-refractory CRC also suggested that patients with p.G13D-mutated tumors showed a trend toward a higher RR than other KRAS-mutated tumors. Patients with KRAS codon p.G13D mutations who received cetuximab experienced longer PFS and OS compared with BSC alone. In contrast, patients with other KRAS mutations did not appear to benefit from cetuximab. Furthermore, benefit from the addition of cetuximab to first-line chemotherapy in patients with KRAS p.G13D mutations has also been suggested in a pooled analysis of the CRYSTAL and OPUS studies[71]. Taken together, these data suggest that the use of cetuximab may affect prolonged survival in patients with KRAS p.G13D mutations receiving first-line chemotherapy and those with chemotherapy-refractory metastatic colon cancer.
The association between KRAS codons 61 and 146 mutations and clinical outcomes in mCRC patients treated with cetuximab has also been investigated[72,73]. It was reported that patients with mCRC that harbors KRAS mutations in codons 61 and 146 have a shorter PFS compared to patients with wild-type KRAS and demonstrate resistance to anti-EGFR therapy[73]. In a prospective-retrospective biomarker analysis of the PRIME study, investigators found that not only KRAS mutations (mutation at codons 12 or 13) are predictive of treatment resistance to EGFR therapy, but RAS mutations (KRAS mutation at codons 61, 117 or 146, NRAS mutation at codons 12, 13, 61, 117 or 146, and BRAF mutations), appear to do the same[74]. These analyses suggest that the assessment of other RAS mutations might help optimize the selection of candidate patients for anti-EGFR mAb therapy.
However, our understanding of the biology of KRAS wild-type/mutated genotype and response to anti-EGFR therapy is far from complete. This is underscored by the fact that approximately 40%-60% of mCRC patients with wild-type KRAS status fail to respond to anti-EGFR therapy[75]. Moreover, mCRC patients with responsive KRAS wild-type tumors inevitably acquire resistance to anti-EGFR therapy and experience tumor progression[76]. A lot of ground remains to be uncovered to clarify the molecular mechanisms that contribute to anti-EGFR therapy resistance/sensitivity, in order that patients can be identified for personalized targeted therapy based on specific genotypes.
BRAF, a component of the RAS/RAF/MEK/ERK/MAPK pathway[66], is thought to function as a downstream effector of KRAS. The BRAF mutation has been identified in 10%-15% of CRC patients[14,77,78]. The most common BRAF mutation in tumors is the V600E mutation, which accounts for 90% of all BRAF mutations in CRC. There is an inverse relationship with KRAS mutation results, with the V600E BRAF mutation seen only in KRAS wild-type tumors[14,73,78]. There is a high concordance in BRAF wild-type status between primary and metastatic tumors, but the level of concordance is lower when the primary tumor harbors a BRAF mutation. BRAF mutation has been shown to be associated with high grade, right sided tumors, female gender, older age and microsatellite instability high (MSI-H) tumors[53,77,79]. It also has been linked to poor survival in advanced CRC independent of therapy[80].
Recently, a series of studies confirmed the potential adverse prognostic impact of BRAF mutations. Yokota et al[81] identified BRAF V600E mutation as an independent prognostic factor for survival in a representative cohort of 229 patients with mCRC. In this study, BRAF mutation was associated with a significantly higher risk of dying from cancer-related causes. This finding is consistent with those of other studies in patients at all disease stages[14,82,83]. In KRAS wild-type patients, BRAF-mutated individuals had a worse outcome in terms of PFS and OS. Furthermore, BRAF is a negative prognostic factor for OS, especially in patients with MSI low (MSI-L) and stable (MSI-S) tumors.
In the CRYSTAL-OPUS pooled analysis, patients whose tumors harbored BRAF mutations had worse PFS and OS compared with those who had both KRAS and BRAF wild-type tumors, independent of treatment with cetuximab[84]. These data are consistent with the biomarker analysis of the CAIRO-2 trial[57,85]. This study investigated a large series of patients with mCRC treated with chemotherapy and bevacizumab with or without cetuximab in a subgroup of 520 patients. BRAF mutations were detected in 45 (8.7%) tumors and was mutually exclusive of KRAS mutations, as reported previously. Patients with BRAF-mutated tumors had a statistically significantly worse PFS and OS compared to patients with wild-type BRAF tumors in both arms of the CAIRO2 trial, however, the RR in the two treatment groups did not differ significantly. The authors concluded that BRAF mutations are not restricted to the outcome of cetuximab treatment[85]. These findings further support the hypothesis that BRAF mutations are negative prognostic biomarkers.
Several retrospective studies have suggested that the occurrence of BRAF V600E mutations accounts for resistance to both cetuximab and panitumumab, but full validation of this association has not been achieved. Di Nicolantonio et al[14] retrospectively examined tumors from 113 patients who had received either cetuximab or panitumumab in a second or successive line chemotherapy regimen. None of the BRAF-mutated patients responded to cetuximab or panitumumab, and none of the responders carried BRAF mutations. BRAF-mutated patients had significantly shorter PFS and OS than wild-type patients. De Roock et al[72] reported 4.7% (36 of 761) of BRAF mutations in a retrospective pooled study of chemorefractory patients from the European Consortium, and patients with BRAF mutations had a significantly lower RR (8.3% vs 38% for wild-type; OR, 0.15; P = 0.0012), shorter PFS (median, 8 wk vs 26 wk for wild-type; HR = 3.74, P < 0.0001) and OS (median, 26 wk vs 54 wk for wild-type; HR = 3.03, P < 0.0001) compared with BRAF wild-type patients. Recently, in the phase III PICCOLO trial[86], designed to evaluate the role of panitumumab combined with irinotecan as second or subsequent line therapy for prospectively tested KRAS wild-type advanced CRC, patients with tumors bearing a BRAF mutation (13.6%) had a poor prognosis and panitumumab had an adverse effect on survival in this subgroup. These results suggest that wild-type BRAF is required for response to anti-EGFR mAb in mCRC. Similarly, Souglakos et al[77] assessed the predictive value of BRAF mutations in 100 patients treated with cetuximab, including 8 in the first line, 37 in the second, and 55 in the third or higher, always in combination with chemotherapy. No patient with BRAF mutations responded to cetuximab. Patients with BRAF mutations also had a shorter PFS, regardless of whether cetuximab was administered in the second, third or higher lines.
However, unlike KRAS mutations, the negative predictive value of BRAF mutations to anti-EGFR therapies in the first-line treatment has not been demonstrated[57,64,84,87]. In the pooled analysis of OPUS and CRYSTAL, patients with BRAF mutations seemed to benefit from the addition of cetuximab to first-line chemotherapy with an increase in OS and a doubling of PFS, although these findings did not reach statistical significance, most likely due to the low BRAF mutation frequency[84]. This result raises the possibility that the addition of a biological agent might be effective for disease control, at least as first-line chemotherapy, in patients with wild-type KRAS and mutant BRAF. These differences were not statistically significant due to the limited number of patients in this group.
The association between BRAF mutations and the efficacy of anti-EGFR therapy remains controversial, but its significant negative prognostic value has been established. Even if the BRAF mutation has been shown to be predictive, its low prevalence suggests that it may have limited utility in selecting patients for anti-EGFR therapy in clinical practice. The novel strategy of targeting BRAF kinase is warranted for further treatment of patients with BRAF mutations to improve their poor survival.
In addition to KRAS and BRAF, activation of the PI3K signaling pathway can also be oncogenically deregulated either by activating mutations in the PIK3CA p110 subunit or by inactivation of the PTEN phosphatase. Constitutive activation of the PI3K/AKT pathway has been hypothesized to play an important role in the development of a number of human cancers, including colon cancer. Activating mutations in the PIK3CA are described in approximately 10%-20% of unselected CRC patients[48,88-90], mainly in exon 9 or 20. Exons 9 and 20 hotspots exert different biochemical and oncogenic properties. Unlike BRAF mutations, PIK3CA mutations can co-occur with KRAS and BRAF mutations[72,91].
Several studies have suggested that PIK3CA mutations may be associated with resistance to EGFR mAb therapy[52,92-94]. Preclinical data shows that colon cancer cell lines with activating PIK3CA mutations were more resistant to cetuximab than PIK3CA wild-type cell lines. Based on these preclinical data, several retrospective studies have evaluated the predictive value of PIK3CA mutations in the clinic. Initial reports show that PIK3CA mutations are able to predict resistance to anti-EGFR mAbs in unselected mCRC patients, and more importantly in wild-type KRAS patients whose nonresponse to treatment cannot be predicted by KRAS mutations[90,95]. Sartore-Bianchi et al[95] found activating PIK3CA mutations in 15 (13.6%) of 110 patients treated with cetuximab or panitumumab-based regimens, but none of the PIK3CA mutated patients achieved a response to anti-EGFR mAbs, compared with a RR of 23% in 95 patients with wild-type PIK3CA (P = 0.0337). Wu et al[92] conducted a systematic review and included eight studies which reported survival outcome in 839 mCRC patients. They found that PIK3CA mutations were significantly associated with poorer PFS in unselected patients, and observed a worse OS in KRAS wild-type patients with PIK3CA mutations. However, the clinical data regarding PIK3CA mutations and response to EGFR mAbs are conflicting. A study by De Roock et al[72] found that PIK3CA mutations in exon 9 were more common (10% of all samples), but only mutations in exon 20 of PIK3CA (3% of all samples) were statistically associated with resistance to cetuximab-based therapy. Importantly, these mutations were also associated with a negative effect on PFS and OS. A meta-analysis by Mao et al[93] recently showed that PIK3CA mutations, in particular in exon 20, were likely to be related to the prognosis of KRAS wild-type mCRC patients treated with anti-EGFR mAbs. The predictive power of exon 20 mutations was also greater than that of any exon mutations and exon 9 mutations. These findings suggest that exon 20 and exon 9 mutations may differ in their power of predicting the prognosis of mCRC patients. If KRAS is unmutated, assessing the PIK3CA exon 20 mutations provides additional information on patient outcome.
The predictive value of PIK3CA mutation status has been demonstrated, however, the prognostic significance of PIK3CA mutations in CRC remains unclear. A number of previous studies have examined the prognostic role of PIK3CA mutations in CRC. Recent data suggest that the presence of PIK3CA mutations predicts poor prognosis for early stage CRC patients and mCRC patients[95,96]. Patients with PIK3CA mutations were more likely to experience local recurrences than patients without mutations[96]. In a study of 586 patients by Barault et al[48], it was found that mutations of at least one gene among KRAS, BRAF and PIK3CA were associated with a lower 3-year survival rate. Kato and coworkers carried out an analysis of 158 CRC tissue samples and identified PIK3CA mutations as the only independent and significant prognostic factor for worse RFS in stage II/III CRC patients[97]. These results are in contrast with those observed in the metastatic setting. Cappuzzo et al[98] described a PIK3CA mutation in 17.7% (14/85) of cetuximab-treated mCRC patients, but found no difference in overall response rate (ORR), time to progression (TTP) and OS compared to the wild-type population. Liao et al[99] analyzed PIK3CA pyrosequencing in 1267 CRC patients, and PIK3CA mutations were detected in 189 (16%) of 1170 cases. The results showed that concomitant PIK3CA mutations of both exons 9 and 20 were associated with a poorer prognosis. In contrast, neither PIK3CA exon 9 mutation nor exon 20 mutation alone appeared to have substantial prognostic influence.
Taken together, these findings are not uniform and there are contradictory reports, thus it is not anticipated that in the short-term future PIK3CA mutation testing will be performed in routine clinical practice to determine eligibility for anti-EGFR antibody therapy. It is also estimated that only 3%-10% of patients who are in the KRAS wild-type group will have a PIK3CA mutation, therefore the potential contribution of this mutation for individualized treatment of CRC will be limited. Thus, further evidence from large randomized clinical trials and standardization of analysis will be required to establish a role for these genetic markers in mCRC treatment.
PTEN is the only tumor suppressor gene involved in the PI3K-AKT-mTOR pathway. It has been shown that inactivation of PTEN phosphatase deregulates the PI3K pathway. PTEN loss is observed in 20%-40% of CRC tumors[94,100], and it has been found to co-occur with KRAS, BRAF and PIK3CA mutations[91,101]. PTEN expression shows only approximately 60% concordance between primary tumor and distant metastases[40,94]. Loss of PTEN expression is associated with aggressive CRC and lack of benefit with cetuximab in patients with chemotherapy-refractory mCRC. It may provide valuable prognostic and predictive information to aid treatment strategies for patients[94].
The prognostic role of PTEN in CRC is still under investigation, and inconclusive results have been reported. In a retrospective analysis of archival tumor tissue from 173 patients with mCRC, loss of PTEN expression (19.9% cases) detected by IHC was associated with inferior OS in a multivariate analysis (HR = 1.9, 95%CI: 1.1-3.2, P = 0.026)[100].
PTEN also shows promise as a predictive marker for wild-type KRAS patients treated with an anti-EGFR-based regimen[102,103]. Wang et al[102] analyzed PTEN expression in 852 mCRC patients treated with anti-EGFR mAbs, and loss of PTEN expression was detected in 242 (28.4%) patients. Anti-EGFR mAb therapy resulted in improved PFS and OS in patients unselected by KRAS mutation with normal PTEN expression over loss of PTEN expression. Better PFS and OS were observed in wild-type KRAS patients with normal PTEN expression vs loss of expression. Razis et al[103] reported that normal PTEN protein expression was associated with a higher RR and longer TTP in patients treated with cetuximab-based therapy, despite a 50% RR observed in patients who had lost PTEN protein expression. These data showed that loss of PTEN expression is a potential biomarker for resistance to anti-EGFR mAb therapy, particularly in mCRC patients with KRAS wild-type tumors. Interestingly, preserved PTEN expression in metastatic samples was predictive of response to cetuximab, while this was not observed in primary tumor tissue with preserved PTEN expression. Therefore, these data are limited and should be considered exploratory. The value of PTEN as a predictive or prognostic marker in mCRC cannot be established yet.
In contrast to gastric and breast cancer, human epidermal growth factor receptor 2 (HER2) protein overexpression and HER2 gene amplification are relatively rare in CRC. Some studies have shown that HER2 gene amplification was significantly related to resistance to cetuximab or panitumumab and was associated with a significantly worse PFS and a trend towards a worse OS[104-106]. However, other studies have not found a predictive or prognostic role for HER2[85,107]. Recently, data from a retrospective study have suggested that HER2 status detected by FISH might represent an additional useful marker for the identification of advanced CRC patients who may benefit from anti-EGFR targeted therapies[105,106]. A total of 407 chemorefractory mCRC patients treated with cetuximab alone or in combination with irinotecan were evaluated and KRAS and BRAF mutations were assessed. The status of the HER2 gene was evaluated in 288 cases. Interestingly, HER2 gene-positive patients had a significantly higher RR, longer PFS and OS compared with HER2 gene-negative patients, but when cases were stratified according to KRAS and BRAF mutations, no significant differences in RR, PFS and OS were observed between HER2-positive and negative cases. In conclusion, the interplay between EGFR and HER2 requires further investigation for future best-tailored treatments.
MET, the hepatocyte growth factor receptor, is a receptor tyrosine kinase (RTK) involved in cellular proliferation and apoptosis. The activation of MET may lead to the activation of pathways downstream of RAS, such as Raf/MEK/MAPK and the PI3K/protein kinase B pathway (PKB). In addition, MET is able to directly activate the PI3K/PKB pathway in a RAS-independent manner[108]. Several preclinical findings suggest that MET can interfere with anti-EGFR strategies. Inno et al[109] recently reported that compared with low/normal expression, c-Met overexpression significantly correlated with shorter median PFS and median OS in 73 patients with mCRC treated with cetuximab-containing regimens. Cappuzzo et al[29] also assessed MET at the genomic level using FISH in 85 EGFR FISH-positive mCRC patients treated with cetuximab. Both patients with MET amplification responded to cetuximab therapy, although the number of patients was too low to draw any conclusion.
Insulin-like growth factor receptor 1 (IGF1R) is also a transmembrane RTK implicated in promoting oncogenic transformation, growth and survival of cancer cells. IGF1R is overexpressed in 50%-90% of CRCs[110], and preclinical studies suggest that this target results in upregulation in the majority of CRC patients, poor prognosis and resistance to anti-EGFR strategies[111].
TP53 is a tumor-suppressor gene located on chromosome 17p, and mutations in this gene occur in about half of CRCs. A large number of studies have described the effects of genetic TP53 alterations on progression and outcome of CRC, and the results are heterogeneous and conflicting. Most studies which showed an association between TP53 alterations and worse outcome employed IHC and the remainder employed DNA analysis. Therefore, it is likely that activation of the EGFR pathway will contribute to cancer and anti-EGFR antibodies will be efficient in tumors only if TP53 is inactivated. Based on these observations, Oden-Gangloff et al[112] evaluated the combined impact of KRAS and TP53 status on clinical outcome in 64 mCRC patients treated with cetuximab-based chemotherapy, and suggested that TP53 mutations are predictive of cetuximab sensitivity.
In conclusion, these data suggest that TP53 genotyping could have an additional value in mCRC patients without KRAS mutations to optimize the selection of patients who could benefit from anti-EGFR therapies. The clinical relevance of these results should be confirmed in larger mCRC series.
Angiogenesis has become a major target in CRC therapy. A variety of anti-angiogenesis approaches have been evaluated for the treatment of mCRC. Bevacizumab, a humanized monoclonal antibody against vascular endothelial growth factor A (VEGF-A), is approved for first-line treatment of mCRC. Novel antiangiogenic drugs, such as regorafenib (a novel tyrosine kinase inhibitor targeting VEGFR, PDGFR, FGFR, RET, KIT and TIE2) and aflibercept (a VEGF trap), have also been licensed by the United States Food and Drug Administration based on trials showing modest improvements in OS[113,114]. However, despite the increasing use of various antiangiogenic drugs and intense research efforts, there is a lack of evidence for validated biomarkers in terms of response to antiangiogenic therapy. Several markers that have appeared promising in preclinical models have failed as predictors of response in human trials (Table 2)[115-117]. To date, no biomarkers have emerged that are capable of predicting the efficacy of these agents.
| Key biomarkers evaluated |
| KRAS mutational status |
| BRAF mutational status |
| p53 mutational status |
| VEGF and VEGFR-2 (KDR) gene expression |
| VEGF A- to VEGF-D, VEGFR-1, and VEGFR-2 protein expression |
| CD31 expression |
| Neuropilin expression |
| Stromal thrombospondin-2 expression |
| Microvessel density |
| Plasma VEGF levels |
Several recent studies on the identification of predictive biomarkers for bevacizumab have been performed. In the pivotal AVF2107 study of bevacizumab added to chemotherapy in the first-line setting of advanced CRC, plasma VEGF levels, primary tumor tissue VEGF expression, microvessel density and genotypic characteristics of the malignant cells such as KRAS, BRAF, TP53 mutations, and TP53 overexpression were evaluated, but none had predictive value for bevacizumab activity. These findings were recently confirmed in the MAX trial, in which the KRAS and BRAF mutation status failed to predict benefit with bevacizumab[118-120]. In this study, the expression levels of VEGF family members A through D and VEGF receptors, VEGFR-1 and VEGFR-2, were also analyzed using IHC, and the results showed that VEGF-D expression was a predictor of response to bevacizumab treatment. For patients treated with bevacizumab, low VEGF-D expression was predictive of a significantly longer PFS and OS interval than those in patients with high levels of VEGF-D expression. In the NO16966 trial[119], exploratory analyses found that high CD31, high VEGF-A, and low EGFR-2 expression levels were correlated with a longer duration of response, and high levels of neuropilin and placental growth factor were associated with less benefit from bevacizumab. However, these results are considered exploratory and need to be confirmed in additional clinical trials.
Blood-based biomarkers have, until now, produced mixed results. Several studies have demonstrated that plasma VEGF-A is a prognostic biomarker in CRC, but it is unable to predict response to antiangiogenic treatment in mCRC[121,122]. A retrospective analysis of 1816 patients with colon, renal cell, and lung cancer found that plasma VEGF levels were not predictive of benefit from bevacizumab[123]. However, an association between plasma VEGF and benefit from bevacizumab treatment was observed in a breast cancer trial[124]. Further prospective studies are underway to validate the value of plasma VEGF-A in clinical practice. VEGF polymorphisms are also potentially promising biomarkers, however, it is not currently possible to personalize treatment with antiangiogenic therapies[125].
More recently, preclinical data supporting the role of fibroblast growth factor receptor (FGFR) and platelet-derived growth factor receptor (PDGFR) signaling in angiogenesis have been reported. Inhibition of these pathways holds potential therapeutic benefit for cancer patients[126]. In addition, one or both of these pathways have been associated with resistance to agents targeting the EGFR and VEGF[127]. Some studies have elucidated the role of FGFR and PDGFR in colon cancer angiogenesis. However, only a few studies have analyzed the clinical implications of FGFR/PDGFR expression in CRC. Wehler et al[128] in a series of 99 human colorectal carcinomas, reported that coexpression of PDGFRα/β observed in 57% of tumor samples, was significantly associated with lymphatic metastasis (P = 0.007) and advanced tumor stage (P = 0.03). Schimanski et al[129] reported that specific receptor tyrosine kinases (TK) were overexpressed in KRAS-mutated CRC. In a study by Nakamura et al[130], patients with high PDGF-BB expression had a significantly poorer survival rate than those with low PDGF-BB expression. A multivariate analysis also demonstrated that PDGFR expression was an independent prognostic factor. Sato et al[131] reported that overexpression of the FGFR1 gene leads to liver metastasis in CRC. Matsuda et al[132] also found overexpression of the FGFR2, both FGFR2IIIc and FGFR2IIIb, in colorectal carcinomas which tended to correlate with distant metastasis. On the other hand, FGFR2IIIb expression in colorectal carcinomas did not correlate with survival or metastasis[133]. It was also found[134] that in colorectal carcinoma cases, expression levels of FGFR2IIIc in tumor cells were correlated with advanced carcinogenesis stages. Furthermore, FGFR2IIIc expression correlated with metastasis and poor prognosis of colorectal carcinomas, which suggested that FGFR2IIIc may have a potential use in colorectal carcinoma therapy. A number of agents that target FGF and/or PDGF signaling are now in development for the treatment of mCRC. Potential predictive biomarkers for these pathways are being investigated, but none have been validated for clinical use. Whether this could translate into a higher likelihood of responding to PDGFR/FGFR targeted agents is a matter of speculation.
Hypertension is a common adverse effect of anti-VEGF therapy. The development of hypertension due to anti-VEGF treatment has also been evaluated as a predictive biomarker. An increase in blood pressure may reflect successful inhibition of the VEGF pathway. However, the role of hypertension in predicting responsiveness to antiangiogenic drugs is controversial. In the AVF2107 study, the development of hypertension predicted better PFS (HR = 0.55, P = 0.0008) and better OS (HR = 0.43, P = 0.0001), but this was not confirmed by other studies[135]. The role of hypertension as a predictive biomarker requires further evaluation, particularly as it is standard practice to treat hypertension as soon as it develops[136].
Epigenetics describe the changes in phenotype or gene expression that do not involve DNA sequence changes. CRC is considered a genetic disease with the histologic progression of carcinogenesis characterized by sequential genetic and epigenetic alterations[137]. Epigenetic instability in CRC is manifested in a variety of ways including hypermethylation of gene promoters that contain CpG islands and global DNA hypomethylation. The role of epigenetics in CRC development and pathogenesis is beginning to be defined. Retrospective studies have proposed candidate markers, such as CpG island methylation (CIMP), which may predict poor outcome for CRC patients after fluorouracil treatment[138]. However, there are conflicting results and studies are required to determine the reproducibility of the data[139]. Promoter CpG island methylation of the Werner syndrome gene[140] and the UDP-glucuronosyl-transferase gene, UGT1A1[141], have been reported to influence the effects of and response to the topoisomerase inhibitor, irinotecan, with these studies being directly related to silencing of genes involved in the mechanism of action of this drug. However, the data are not currently robust enough to recommend its clinical use[142,143].
Epigenetic changes in CRC are also potential markers for the early detection of CRC and prediction of prognosis. Several publications report a prognostic role for promoter CIMP markers, such as p16INK4A, p14ARF, MGMT, HPP1, HLTF, and ID4, but their effects seem to be dependent on the presence of other methylated markers or adjuvant treatment[139]. A prognostic role was also suggested for CIMP, and a worse prognosis for patients with CIMP CRCs was observed in most studies, although conflicting results have also been reported[140]. These examples of the potential prognostic use of alterations in DNA methylation highlight the need for validation of their clinical utility in observational, population-based studies to assess the natural course of the disease.
Despite these examples and other studies of predictive and prognostic epigenetic markers in CRC, none have yet been developed to the point of clinical utility. Continued efforts to investigate these molecular mechanisms will allow for a better understanding of the role of epigenetic alterations in CRC and will lead to the translation of these insights into the clinical arena.
Currently, the treatment of advanced CRC varies and oncologists face complicated decisions in the selection of the most appropriate treatment options for their patients. Predictive and prognostic biomarkers can facilitate clinical decision-making and are becoming increasingly important with the development of targeted therapies for advanced CRC. The identification of molecular biomarkers that have predictive and/or prognostic significance in CRC is essential to improve anti-cancer treatments and patient outcome[144]. Several molecular biomarkers have been studied over the past two decades and encouraging improvements have been achieved. However, the results of published studies have often been conflicting and several drawbacks affect the reliability of conclusions[145]. First, most published studies were retrospective analyses of a single marker or included a small sample size. These study designs are unlikely to accurately predict disease progression with sufficient resolution and reproducibility. Second, data analysis and interpretation still remain challenging, although many advances have been made in technologies for profiling and in decreasing the requirements of the input material. The data from current studies usually lack definition, adequate validation, and cannot be used in clinical practice for decision-making. Furthermore, the lack of methodology standardization involved in the detection of biomarkers, the lack of comprehensive analysis of a particular molecular pathway, and incomplete analysis of biomarkers have all contributed to the frustration associated with biomarker validation. Therefore, to date, only KRAS gene has entered routine clinical practice as a predictive marker of response to EGFR-targeted therapies in advanced CRC.
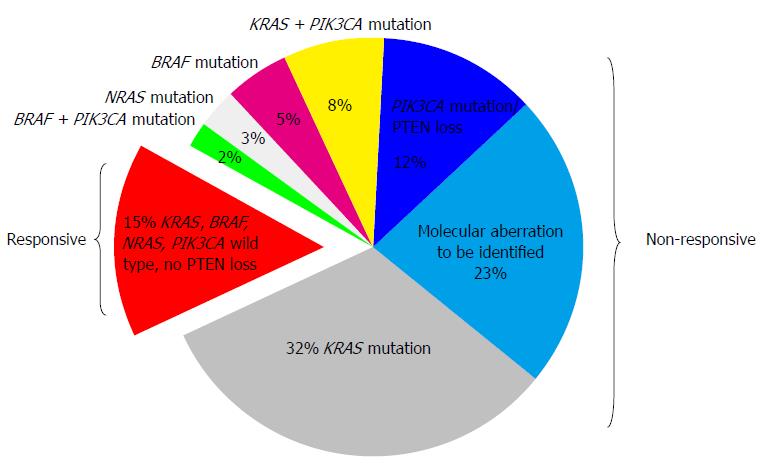
A number of comprehensive biomarker-driven studies are currently underway. BRAF V600E mutation is prognostic of patient outcome with respect to survival, but not clearly predictive of treatment effects with anti-EGFR agents in patients with mCRC. The low prevalence of such mutations makes it difficult to evaluate these mutations as predictive biomarkers in clinical practice. The predictive and prognostic value of PIK3CA mutation, PTEN deletion and TP53 mutation is presently under evaluation, but clinicians are currently unable to use these data in clinical practice for decision-making. In the future, NRAS, PIK3CA and PTEN status may be useful when combined with KRAS and BRAF mutation analysis to predict which mCRC patients will benefit from anti-EGFR therapy (Figure 1). The identification of a biomarker to predict response to anti-VEGF agents is lacking, and further data are required from large well designed prospective studies to understand the biological processes underlying response and/or resistance. Novel prospective randomized controlled trials are needed to determine the role of various putative molecular markers, and hopefully this will facilitate the development of personalized therapy based on the molecular profile of CRC.
In addition to these molecular markers, many patient-related factors may also influence response to targeted therapy, including age, sex, tumor subtype, disease stage, comorbid diseases, overall PS, pharmacokinetic, pharmacodynamic and pharmacogenetic factors. These factors should be considered as important predictive and prognostic biomarkers in CRC.
In the future, it is anticipated that new biomarkers will be developed that can further personalize the treatment of this important human cancer. In the era of targeted therapies, it is further anticipated that new small molecule drugs that target specific gene mutations (for example, BRAF inhibitors) and genetic translocations will be developed in association with specific biomarker tests that are linked to drug response and patient eligibility for treatment.
P- Reviewers: Clarke SJ, De Petris L, Kato J, Li QQ S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Wang N, Sun TT, Zheng RS, Zhang SW, Chen WQ. An Analysis of Incidence and Mortality of Colorectal Cancer in China, 2009. Zhongguo Zhongliu. 2013;22:516-520. |
| 2. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10453] [Article Influence: 696.9] [Reference Citation Analysis (0)] |
| 3. | De Mattos-Arruda L, Dienstmann R, Tabernero J. Development of molecular biomarkers in individualized treatment of colorectal cancer. Clin Colorectal Cancer. 2011;10:279-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 385] [Cited by in RCA: 369] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 5. | Winer E, Gralow J, Diller L, Karlan B, Loehrer P, Pierce L, Demetri G, Ganz P, Kramer B, Kris M. Clinical cancer advances 2008: major research advances in cancer treatment, prevention, and screening--a report from the American Society of Clinical Oncology. J Clin Oncol. 2009;27:812-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12:5268-5272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 649] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 7. | Harding J, Burtness B. Cetuximab: an epidermal growth factor receptor chemeric human-murine monoclonal antibody. Drugs Today (Barc). 2005;41:107-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 188] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 8. | Lockhart AC, Berlin JD. The epidermal growth factor receptor as a target for colorectal cancer therapy. Semin Oncol. 2005;32:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Ng K, Zhu AX. Targeting the epidermal growth factor receptor in metastatic colorectal cancer. Crit Rev Oncol Hematol. 2008;65:8-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:71-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 11. | Vincenzi B, Zoccoli A, Pantano F, Venditti O, Galluzzo S. Cetuximab: from bench to bedside. Curr Cancer Drug Targets. 2010;10:80-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Giusti RM, Cohen MH, Keegan P, Pazdur R. FDA review of a panitumumab (Vectibix) clinical trial for first-line treatment of metastatic colorectal cancer. Oncologist. 2009;14:284-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643-2648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 621] [Cited by in RCA: 637] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 14. | Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705-5712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1211] [Cited by in RCA: 1242] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 15. | Spindler KL, Lindebjerg J, Nielsen JN, Olsen DA, Bisgård C, Brandslund I, Jakobsen A. Epidermal growth factor receptor analyses in colorectal cancer: a comparison of methods. Int J Oncol. 2006;29:1159-1165. [PubMed] |
| 16. | Moroni M, Sartore-Bianchi A, Benvenuti S, Artale S, Bardelli A, Siena S. Somatic mutation of EGFR catalytic domain and treatment with gefitinib in colorectal cancer. Ann Oncol. 2005;16:1848-1849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Scartozzi M, Bearzi I, Pierantoni C, Mandolesi A, Loupakis F, Zaniboni A, Catalano V, Quadri A, Zorzi F, Berardi R. Nuclear factor-kB tumor expression predicts response and survival in irinotecan-refractory metastatic colorectal cancer treated with cetuximab-irinotecan therapy. J Clin Oncol. 2007;25:3930-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308-1324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 413] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 19. | Rodríguez J, Viúdez A, Ponz-Sarvisé M, Gil-Aldea I, Chopitea A, García-Foncillas J, Gil-Bazo I. Improving disease control in advanced colorectal cancer: Panitumumab and cetuximab. Crit Rev Oncol Hematol. 2010;74:193-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1471] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 21. | Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1306] [Cited by in RCA: 1283] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 22. | Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 821] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 23. | Lenz HJ, Van Cutsem E, Khambata-Ford S, Mayer RJ, Gold P, Stella P, Mirtsching B, Cohn AL, Pippas AW, Azarnia N. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol. 2006;24:4914-4921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 392] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 24. | Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer. 2001;92:1331-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 25. | Taron M, Ichinose Y, Rosell R, Mok T, Massuti B, Zamora L, Mate JL, Manegold C, Ono M, Queralt C. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878-5885. [PubMed] |
| 26. | Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 250] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 27. | Tsuchihashi Z, Khambata-Ford S, Hanna N, Jänne PA. Responsiveness to cetuximab without mutations in EGFR. N Engl J Med. 2005;353:208-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 28. | Personeni N, Fieuws S, Piessevaux H, De Hertogh G, De Schutter J, Biesmans B, De Roock W, Capoen A, Debiec-Rychter M, Van Laethem JL. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res. 2008;14:5869-5876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 29. | Cappuzzo F, Finocchiaro G, Rossi E, Jänne PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F, Pressiani T. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19:717-723. [PubMed] |
| 30. | Yang ZY, Shen WX, Hu XF, Zheng DY, Wu XY, Huang YF, Chen JZ, Mao C, Tang JL. EGFR gene copy number as a predictive biomarker for the treatment of metastatic colorectal cancer with anti-EGFR monoclonal antibodies: a meta-analysis. J Hematol Oncol. 2012;5:52. [PubMed] |
| 31. | Jiang Z, Li C, Li F, Wang X. EGFR gene copy number as a prognostic marker in colorectal cancer patients treated with cetuximab or panitumumab: a systematic review and meta analysis. PLoS One. 2013;8:e56205. [PubMed] |
| 32. | Ooi A, Takehana T, Li X, Suzuki S, Kunitomo K, Iino H, Fujii H, Takeda Y, Dobashi Y. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol. 2004;17:895-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Adams R, Fisher D, Farragher S, Scott A, Smith C, James M, Cheadle J, Nichols L, Meade AM, Kaplan RS. Use of epiregulin (EREG) and amphiregulin (AREG) gene expression to predict response to cetuximab therapy in combination with oxaliplatin (Ox) and 5FU in first-line treatment of advanced colorectal cancer (aCRC). Proceedings of ASCO Annual Meeting; 2012 May 30-June 3; McCormick Place, Chicago, Illinois. J Clin Oncol. 2012;Abstract 3516. |
| 34. | Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230-3237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 897] [Cited by in RCA: 893] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 35. | Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, Fieuws S, Vandesompele J, Peeters M, Van Laethem JL. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27:5068-5074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 277] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 36. | de Reyniès A, Boige V, Milano G, Faivre J, Laurent-Puig P. KRAS mutation signature in colorectal tumors significantly overlaps with the cetuximab response signature. J Clin Oncol. 2008;26:2228-2230; author reply 2230-2231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Tabernero J, Cervantes A, Rivera F, Martinelli E, Rojo F, von Heydebreck A, Macarulla T, Rodriguez-Braun E, Eugenia Vega-Villegas M, Senger S. Pharmacogenomic and pharmacoproteomic studies of cetuximab in metastatic colorectal cancer: biomarker analysis of a phase I dose-escalation study. J Clin Oncol. 2010;28:1181-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, Tsakalaki E, Trypaki M, Messaritakis I, Stathopoulos E. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ≥ 2 line cetuximab-based therapy of colorectal cancer patients. PLoS One. 2011;6:e15980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 39. | Kuramochi H, Nakajima G, Kaneko Y, Nakamura A, Inoue Y, Okuyama R, Kondo Y, Kanemura T, Hayashi K, Yamamoto M. Amphiregulin and epiregulin mRNA expression in primary colorectal cancer and corresponding liver metastases. Proceedings of ASCO Annual Meeting; 2011 Jun 9; McCormick Place, Chicago, Illinois. J Clin Oncol. 2011;e14023. |
| 40. | Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1275] [Cited by in RCA: 1246] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 41. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] |
| 42. | Artale S, Sartore-Bianchi A, Veronese SM, Gambi V, Sarnataro CS, Gambacorta M, Lauricella C, Siena S. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J Clin Oncol. 2008;26:4217-4219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 43. | Zauber P, Sabbath-Solitare M, Marotta SP, Bishop DT. Molecular changes in the Ki-ras and APC genes in primary colorectal carcinoma and synchronous metastases compared with the findings in accompanying adenomas. Mol Pathol. 2003;56:137-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682-4689. [PubMed] |
| 45. | Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928-932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 46. | Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 546] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 47. | Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692-696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 632] [Cited by in RCA: 666] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 48. | Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM, Lièvre A, Cortet M, Bouvier AM, Rat P. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255-2259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 246] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 49. | Dix BR, Robbins P, Soong R, Jenner D, House AK, Iacopetta BJ. The common molecular genetic alterations in Dukes’ B and C colorectal carcinomas are not short-term prognostic indicators of survival. Int J Cancer. 1994;59:747-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 87] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 50. | Esteller M, González S, Risques RA, Marcuello E, Mangues R, Germà JR, Herman JG, Capellà G, Peinado MA. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001;19:299-304. [PubMed] |
| 51. | Alberts SR, Sargent DJ, Smyrk TC, Shields AF, Chan E, Goldberg RM, Gill S, Kahlenberg MS, Thibodeau SN, Nair S. Adjuvant mFOLFOX6 with or without cetuxiumab (Cmab) in KRAS wild-type (WT) patients (pts) with resected stage III co- lon cancer (CC): Results from NCCTG Intergroup Phase III Trial N0147. Proceedings of ASCO Annual Meeting; 2010 June 22; McCormick Place, Chicago, Illinois. J Clin Oncol. 2010;CRA3507. |
| 52. | Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103-2114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 766] [Cited by in RCA: 763] [Article Influence: 54.5] [Reference Citation Analysis (2)] |
| 53. | Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 917] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 54. | Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, Hantel A. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322-7329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 55. | Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2724] [Cited by in RCA: 2761] [Article Influence: 162.4] [Reference Citation Analysis (0)] |
| 56. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2504] [Cited by in RCA: 2403] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 57. | Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1000] [Cited by in RCA: 999] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 58. | Hecht JR, Mitchell E, Chidiac T, Scroggin C, Hagenstad C, Spigel D, Marshall J, Cohn A, McCollum D, Stella P. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol. 2009;27:672-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 636] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 59. | Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992-3995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1669] [Cited by in RCA: 1699] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 60. | Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1218] [Cited by in RCA: 1240] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 61. | Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2901] [Cited by in RCA: 3122] [Article Influence: 195.1] [Reference Citation Analysis (1)] |
| 62. | Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697-4705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1296] [Cited by in RCA: 1393] [Article Influence: 92.9] [Reference Citation Analysis (0)] |
| 63. | Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, André T, Chan E, Lordick F, Punt CJ. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:4706-4713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 709] [Cited by in RCA: 758] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 64. | Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535-1546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 586] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 65. | Tveit KM, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, Sigurdsson F, Kure E, Ikdahl T, Skovlund E. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol. 2012;30:1755-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 420] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 66. | Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279-286. [PubMed] |
| 67. | Lee CN, Chen HY, Liu HE. Favorable response to erlotinib in a lung adenocarcinoma with both epidermal growth factor receptor exon 19 deletion and K-ras G13D mutations. J Clin Oncol. 2010;28:e111-e112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 68. | De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 626] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 69. | Guerrero S, Casanova I, Farré L, Mazo A, Capellà G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750-6756. [PubMed] |
| 70. | De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 584] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 71. | Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570-3577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 287] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 72. | De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1491] [Cited by in RCA: 1656] [Article Influence: 110.4] [Reference Citation Analysis (1)] |
| 73. | Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715-721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 448] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 74. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1610] [Cited by in RCA: 1730] [Article Influence: 144.2] [Reference Citation Analysis (0)] |
| 75. | Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 533] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 76. | Weickhardt AJ, Tebbutt NC, Mariadason JM. Strategies for overcoming inherent and acquired resistance to EGFR inhibitors by targeting downstream effectors in the RAS/PI3K pathway. Curr Cancer Drug Targets. 2010;10:824-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 77. | Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465-472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 242] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 78. | Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931-5937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 464] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 79. | Vilkin A, Niv Y, Nagasaka T, Morgenstern S, Levi Z, Fireman Z, Fuerst F, Goel A, Boland CR. Microsatellite instability, MLH1 promoter methylation, and BRAF mutation analysis in sporadic colorectal cancers of different ethnic groups in Israel. Cancer. 2009;115:760-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 80. | Rizzo S, Bronte G, Fanale D, Corsini L, Silvestris N, Santini D, Gulotta G, Bazan V, Gebbia N, Fulfaro F. Prognostic vs predictive molecular biomarkers in colorectal cancer: is KRAS and BRAF wild type status required for anti-EGFR therapy. Cancer Treat Rev. 2010;36 Suppl 3:S56-S61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 81. | Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856-862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 319] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 82. | Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396-2402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 83. | Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 633] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 84. | Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M, Celik I, Köhne CH. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 433] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 85. | Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Börger ME, van Cleef PH, van Krieken JH, Punt CJ, Nagtegaal ID. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 86. | Seymour MT, Brown SR, Middleton G, Maughan T, Richman S, Gwyther S, Lowe C, Seligmann JF, Wadsley J, Maisey N. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14:749-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 87. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1314] [Cited by in RCA: 1452] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 88. | Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 327] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 89. | Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, Yan L, Longtine JA, Fuchs CS, Ogino S. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 199] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 90. | Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, Lambrechts D, Van Cutsem E, Tejpar S. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15:3184-3188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 231] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 91. | Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, Martini M, Cipani T, Marrapese G, Mazzucchelli L. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009;4:e7287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 210] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 92. | Wu S, Gan Y, Wang X, Liu J, Li M, Tang Y. PIK3CA mutation is associated with poor survival among patients with metastatic colorectal cancer following anti-EGFR monoclonal antibody therapy: a meta-analysis. J Cancer Res Clin Oncol. 2013;139:891-900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 93. | Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23:1518-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 94. | Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 331] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 95. | Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 595] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 96. | He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, van Velthuysen ML, Zeestraten EC, Nagtegaal ID, van de Velde CJ, Marijnen CA. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15:6956-6962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 97. | Kato S, Iida S, Higuchi T, Ishikawa T, Takagi Y, Yasuno M, Enomoto M, Uetake H, Sugihara K. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer. 2007;121:1771-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 98. | Cappuzzo F, Varella-Garcia M, Finocchiaro G, Skokan M, Gajapathy S, Carnaghi C, Rimassa L, Rossi E, Ligorio C, Di Tommaso L. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 99. | Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257-2268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 214] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 100. | Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924-5930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 525] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 101. | Molinari F, Martin V, Saletti P, De Dosso S, Spitale A, Camponovo A, Bordoni A, Crippa S, Mazzucchelli L, Frattini M. Differing deregulation of EGFR and downstream proteins in primary colorectal cancer and related metastatic sites may be clinically relevant. Br J Cancer. 2009;100:1087-1094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 102. | Wang ZH, Gao QY, Fang JY. Loss of PTEN expression as a predictor of resistance to anti-EGFR monoclonal therapy in metastatic colorectal cancer: evidence from retrospective studies. Cancer Chemother Pharmacol. 2012;69:1647-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 103. | Razis E, Briasoulis E, Vrettou E, Skarlos DV, Papamichael D, Kostopoulos I, Samantas E, Xanthakis I, Bobos M, Galanidi E. Potential value of PTEN in predicting cetuximab response in colorectal cancer: an exploratory study. BMC Cancer. 2008;8:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 104. | Barbara C, Martin V, Molinari F, Molinari F, Landi L, Riva A, Saletti P, Dosso S, Geva R, Tejpar S. Use of HER2 gene amplification to identify patients with metastatic colorectal cancer resistant to anti-EGFR monoclonal antibodies. Presented at the 2012 Gastrointestinal Cancers Symposium; 2010 Jan 28-30; San Francisco, California. J Clin Oncol. 2012;Abst474. |
| 105. | Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, Saletti P, De Dosso S, Spitale A, Tejpar S. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668-675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 106. | Martin V, Sacconi A, Landi L, Riva A, Saletti P, Geva R, Tejpar S, Kalogeras K, Frattini M, Cappuzzo F. An International Consortium in chemorefractory metastatic colorectal cancer patients shows cetuximab efficacy in patients harboring HER2 gene copy number. European Multidisciplinary Cancer Congress; 2011 Sep 23-27; Stockholm, Sweden. Eur J Cancer. 2011;S392 [abstr 6015]. |
| 107. | Troiani T, Zappavigna S, Martinelli E, Addeo SR, Stiuso P, Ciardiello F, Caraglia M. Optimizing treatment of metastatic colorectal cancer patients with anti-EGFR antibodies: overcoming the mechanisms of cancer cell resistance. Expert Opin Biol Ther. 2013;13:241-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 108. | Jiang W, Hiscox S, Matsumoto K, Nakamura T. Hepatocyte growth factor/scatter factor, its molecular, cellular and clinical implications in cancer. Crit Rev Oncol Hematol. 1999;29:209-248. [PubMed] |
| 109. | Inno A, Di Salvatore M, Cenci T, Martini M, Orlandi A, Strippoli A, Ferrara AM, Bagalà C, Cassano A, Larocca LM. Is there a role for IGF1R and c-MET pathways in resistance to cetuximab in metastatic colorectal cancer. Clin Colorectal Cancer. 2011;10:325-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 110. | Koda M, Reszec J, Sulkowska M, Kanczuga-Koda L, Sulkowski S. Expression of the insulin-like growth factor-I receptor and proapoptotic Bax and Bak proteins in human colorectal cancer. Ann N Y Acad Sci. 2004;1030:377-383. [PubMed] |
| 111. | Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62:200-207. [PubMed] |
| 112. | Oden-Gangloff A, Di Fiore F, Bibeau F, Lamy A, Bougeard G, Charbonnier F, Blanchard F, Tougeron D, Ychou M, Boissière F. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer. 2009;100:1330-1335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 113. | Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2194] [Cited by in RCA: 2112] [Article Influence: 176.0] [Reference Citation Analysis (0)] |
| 114. | Van Cutsem E, Tabernero J, Lakomy R, Prenen H, Prausová J, Macarulla T, Ruff P, van Hazel GA, Moiseyenko V, Ferry D. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol. 2012;30:3499-3506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 1024] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 115. | Crawford Y, Kasman I, Yu L, Zhong C, Wu X, Modrusan Z, Kaminker J, Ferrara N. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell. 2009;15:21-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 503] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 116. | Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67:11244-11253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 283] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 117. | Pan Q, Chanthery Y, Liang WC, Stawicki S, Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell. 2007;11:53-67. [PubMed] |
| 118. | Price TJ, Hardingham JE, Lee CK, Weickhardt A, Townsend AR, Wrin JW, Chua A, Shivasami A, Cummins MM, Murone C. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29:2675-2682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 119. | Foernzler D, Delmar P, Kockx M, Cassidy J, Saltz L, Scherer S. Tumor tissue based biomarker analysis in NO16966: A randomized phase III study of first-line bevacizumab in combination with oxaliplatin-based chemotherapy in patients with mCRC [abstract 374]. Presented at the 2010 Gastrointestinal Cancers Symposium; 2010 Jan 22-24; Orlando, FL. . |
| 120. | Weickhardt AJ, Williams D, Lee C, Simes C, Murone C, Wilson K, Cummins M, Asadi K, Price TJ, Mariadason J. Vascular endothelial growth factors (VEGF) and VEGF receptor expression as predictive biomarkers for benefit with bevacizumab in metastatic colorectal cancer (mCRC): Analysis of the phase III MAX study. Proceedings of 2011 ASCO Annual Meeting; Jun 9; Chicago, Illinois. J Clin Oncol. 2011;Abst3531. |
| 121. | Jubb AM, Harris AL. Biomarkers to predict the clinical efficacy of bevacizumab in cancer. Lancet Oncol. 2010;11:1172-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 122. | Jayson GC, de Haas S, Delmar P, Miles DW, Shah MA, Van Cutsem E, Carmeliet P, Hegde P, Wilds N, Scherer S J. Evaluation of plasma VEGFA as a potential predictive pan-tumor biomarker for bevacizumab. Proceedings of 2011 European Multidisciplinary Cancer Congress; 2011 Sep 23-27; Stockholm, Sweden. Eur J Cancer. 2011;S96. |
| 123. | Bernaards C, Hegde P, Chen D, Holmgren E, Zheng M, Jubb AM, Koeppen H, Scherer SJ, Chen DS. Circulating vascular endothelial growth factor (VEGF) as a biomarker for bevacizumab-based therapy in meta-static colorectal, non-small cell lung, and renal cell cancers: Analysis of phase III studies. Proceedings of 2010 ASCO Annual Meeting; Jun 14; Chicago, Illinois. J Clin Oncol. 2010;Abst10519. |
| 124. | Miles DW, de Haas SL, Dirix L, Chan A, Pivot X, Tomczak P, Provencher L, Delmar P, Scherer S. Plasma biomarker analyses in the AVADO phase III randomized study of first-line bevacizumab docetaxel in patients with human epidermal growth factor receptor (HER) 2-negative metastatic breast cancer. Cancer Res. 2010;70:Abstract nr P2-16-04. [DOI] [Full Text] |
| 125. | Koutras AK, Antonacopoulou AG, Eleftheraki AG, Dimitrakopoulos FI, Koumarianou A, Varthalitis I, Fostira F, Sgouros J, Briasoulis E, Bournakis E. Vascular endothelial growth factor polymorphisms and clinical outcome in colorectal cancer patients treated with irinotecan-based chemotherapy and bevacizumab. Pharmacogenomics J. 2012;12:468-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 126. | Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6437] [Cited by in RCA: 6485] [Article Influence: 259.4] [Reference Citation Analysis (0)] |
| 127. | Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 2008;25:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 128. | Wehler TC, Frerichs K, Graf C, Drescher D, Schimanski K, Biesterfeld S, Berger MR, Kanzler S, Junginger T, Galle PR. PDGFRalpha/beta expression correlates with the metastatic behavior of human colorectal cancer: a possible rationale for a molecular targeting strategy. Oncol Rep. 2008;19:697-704. [PubMed] |
| 129. | Schimanski CC, Zimmermann T, Schmidtmann I, Gockel I, Lang H, Galle PR, Moehler M, Berger MR. K-ras mutation status correlates with the expression of VEGFR1, VEGFR2, and PDGFRalpha in colorectal cancer. Int J Colorectal Dis. 2010;25:181-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Nakamura Y, Tanaka F, Yoshikawa Y, Mimori K, Inoue H, Yanaga K, Mori M. PDGF-BB is a novel prognostic factor in colorectal cancer. Ann Surg Oncol. 2008;15:2129-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 131. | Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, Fujii S, Kunisaki C, Shiozawa M, Akaike M. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009;21:211-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 132. | Matsuda Y, Ishiwata T, Yamahatsu K, Kawahara K, Hagio M, Peng WX, Yamamoto T, Nakazawa N, Seya T, Ohaki Y. Overexpressed fibroblast growth factor receptor 2 in the invasive front of colorectal cancer: a potential therapeutic target in colorectal cancer. Cancer Lett. 2011;309:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 133. | Yoshino M, Ishiwata T, Watanabe M, Komine O, Shibuya T, Tokunaga A, Naito Z. Keratinocyte growth factor receptor expression in normal colorectal epithelial cells and differentiated type of colorectal cancer. Oncol Rep. 2005;13:247-252. [PubMed] |
| 134. | Matsuda Y, Hagio M, Seya T, Ishiwata T. Fibroblast growth factor receptor 2 IIIc as a therapeutic target for colorectal cancer cells. Mol Cancer Ther. 2012;11:2010-2020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 135. | Dewdney A, Cunningham D, Barbachano Y, Chau I. Correlation of bevacizumab-induced hypertension and outcome in the BOXER study, a phase II study of capecitabine, oxaliplatin (CAPOX) plus bevacizumab as peri-operative treatment in 45 patients with poor-risk colorectal liver-only metastases unsuitable for upfront resection. Br J Cancer. 2012;106:1718-1721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 136. | Syrigos KN, Karapanagiotou E, Boura P, Manegold C, Harrington K. Bevacizumab-induced hypertension: pathogenesis and management. BioDrugs. 2011;25:159-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 137. | van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol. 2011;29:1382-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 138. | Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 139. | Smits KM, Cleven AH, Weijenberg MP, Hughes LA, Herman JG, de Bruïne AP, van Engeland M. Pharmacoepigenomics in colorectal cancer: a step forward in predicting prognosis and treatment response. Pharmacogenomics. 2008;9:1903-1916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 140. | Agrelo R, Cheng WH, Setien F, Ropero S, Espada J, Fraga MF, Herranz M, Paz MF, Sanchez-Cespedes M, Artiga MJ. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci USA. 2006;103:8822-8827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 141. | Gagnon JF, Bernard O, Villeneuve L, Têtu B, Guillemette C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin Cancer Res. 2006;12:1850-1858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 142. | Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup. Int J Clin Oncol. 2008;13:498-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 60] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 143. | Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104:18654-18659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 418] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 144. | Kahlenberg MS, Sullivan JM, Witmer DD, Petrelli NJ. Molecular prognostics in colorectal cancer. Surg Oncol. 2003;12:173-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 145. | Graziano F, Catalano V, Baldelli AM, Cascinu S. Prognostic biomarkers in resected colorectal cancer: implications for adjuvant chemotherapy. Expert Rev Anticancer Ther. 2001;1:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |