Published online Sep 15, 1996. doi: 10.3748/wjg.v2.i3.161
Revised: July 15, 1996
Accepted: August 13, 1996
Published online: September 15, 1996
AIM: To examine the prevalence of p53 mutations in cases of hepatocellular carcinoma (HCC) in the Chongqing area of China, and the relationship between p53 mutations and the clinicopathological features of HCC, as well as its risk factors.
METHODS: The overexpression and point mutations of tumor suppressor gene p53 in 38 cases of HCC were detected by a sensitive antigen retrieval fluid (ARF) immunohistochemical method, polymerase chain reaction (PCR) a2 restriction fragment length polymorphism (RFLP) analysis, and a single strand conformation polymorphism (SSCP) a2silver staining analysis.
RESULTS: The results showed that 16 of 38 HCCs were positive for P53 protein (42.1%). Seven HCCs (18.4%) had a p53 mutation at codon 249, and 2 other HCCs had point a mutation within exon 7 but not at codon 249. Among 9 cases of HCC which showed genetic mutations, 8 cases (88.9%) were positive for p53 protein. Both p53 overexpression and mutation were significantly related to the degree of HCC tissue differentiation and the presence or absence of metastases. The frequency of p53 mutations was consistent with the high prevalence of HBV infection and moderate aflatoxin B1 (AFB1) exposure in the Chongqing area.
CONCLUSION: Our results suggest that AFB1 acts synergistically with HBV in the generation of p53 mutations. Furthermore, dietary exposure to AFB1 may mainly contribute to the tumor specific mutation at codon 249, while HBV may account for other scattered mutations found in cases of HCC.
- Citation: Wang D, Shi JQ. Overexpression and mutations of tumor suppressor gene p53 in hepatocellular carcinoma. World J Gastroenterol 1996; 2(3): 161-164
- URL: https://www.wjgnet.com/1007-9327/full/v2/i3/161.htm
- DOI: https://dx.doi.org/10.3748/wjg.v2.i3.161
Hepatocellular carcinoma (HCC) is one of the most common cancers in China. A striking correlation exists between the development of HCC and infection with the hepatitis B virus (HBV), as well as exposure to aflatoxin B1 (AFB1). However, the molecular mechanisms underlying these correlations are largely unknown. Tumor suppressor gene p53, located on the short arm of human chromosome 17 (17 p 13.1), is one of the most important and broad spectrum anti-oncogenes, and researchers have documented > 51 types of human tumors that have p53 mutations. Since Bressac[1] and Hsu et al[2] first reported in 1991 that ~ 50% of resected HCCs in China and South Africa had p53 mutations, increasing numbers of similar reports have been published by researchers in various areas of the world. It is very interesting that the presence of p53 mutations in HCC appears to vary geographically[3]. The city of Chongqing is located in southwestern China, an area with a high prevalence of HBV infections, and a moderate exposure to dietary AFB1. However, to date, there has been no report concerning the HCC incidence in that geographic region.
In this study, we used antigen retrieval fluid (ARF) immunohisto-chemistry, PCR, and PCR-SSCP silver staining methods to detect the gene overexpressions and mutations that existed in 38 cases of HCC. Our goal was to demonstrate the prevalence of p53 mutations in cases of HCC in the Chongqing region of China, and investigate the relationship between the p53 alterations and the clinicopathological features of HCC, as well as HCC risk factors.
Between 1992 and 1993, 38 samples of resected HCC tissue (one sample per case) were obtained from the Southwest Hospital affiliated with our college. Thirty-six of the samples were fresh tissue. The tissue specimens were fixed in 10% formalin and embedded in paraffin; after which, 5 μm thick sections were cut for use in our study. When based on their WHO classification, the tissue specimens represented the following types of HCC: trabecular (n = 15), pseudoglandular (n = 16), and compact (n = 7). Histological grading of the specimens based on Edmondson and Steiner’s standard showed: grade I, 3 cases; grade , 7 cases; grade III, 15 cases; grade IV, 13 cases. The tumors could be divided into the following three groups depending on their diameter: 3 cases, ≤ 3 cm; 5 cases, 3 cm-5 cm; 30 cases, ≥ 5 cm. Fourteen cases had intrahepatic metastases at the time of operation.
High-molecular weight DNA was prepared from 36 fresh tissue samples, 2 paraffin-embedded blocks, and 2 samples of normal lymphocytes using promase K and phenol-chloroform methods as described in “Molecular Cloning: A laboratory Manual.”
Monoclonal antibody Pab1801 donated by Dr. L. Crawford of ICRF, United Kingdom, and polyclonal antiserum CM-1, donated by Dr. D.P. Lane, Dundee, United Kingdom, were used to detect p53 protein expression. An ABC kit was purchased from the Dako Company (Demark). Immunohistochemical staining with enhancement was performed using the method described by Vendenberg et al[4,5].
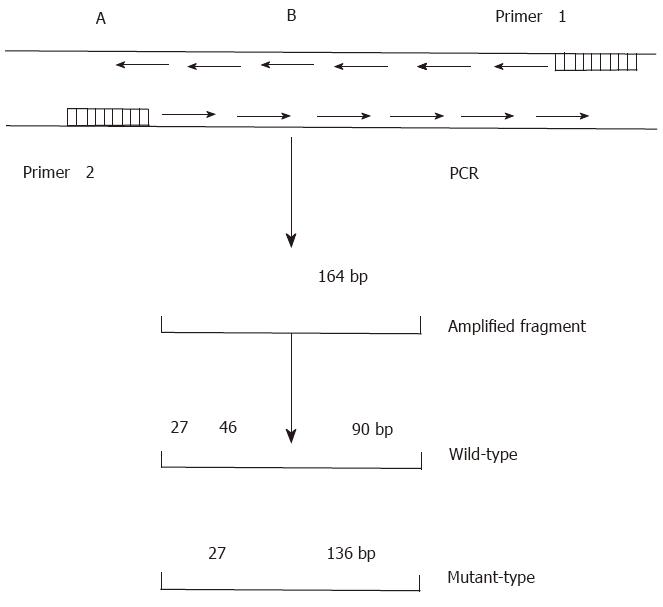
The DNA sequence of p53 exon 7 was amplified by PCR (Figure 1) using primers with the following nucleotide sequences:
Primer 1: 5’CCCAAGGCGCACTGACCTCA3’;
Primer 2: 5’GCTCCTGACCTGGAGTCTTC3’.
PCR was performed according to a standard protocol. Briefly, each PCR reaction was performed in a final volume of 40 μL, which contained 1.8 μL of target DNA, 28 μL ddH2O, 3.2 μL of 2.5 mmol/L dNTP, 2 μL of 20 pmol/L primers, and 4 μL of 10 × Taq DNA polymerase buffer. The samples and reagents were heated for 8 min at 93 °C to denature the DNA. Next, 1 μL (2.2 U) of Taq DNA polymerase was added to each sample, which was then heated for 35 s at 94 °C for denaturation, followed by 40 s at 54 °Cfor annealing, and 60 s at 72 °C for extension. After a total of 35 rounds, each sample was maintained at 72 °C for 10 min. The final DNA product was purified with phenol/chloroform, precipitated with ethanol, dissolved with TE buffer, and then digested with Hae III for 12-24 h. The digests were separated by electrophoresis on an 8% polyacrylamide gel.
A 5 μL sample DNA was mixed with a loading solution containing 950 mM deionized formamide, 10 mmol/L EDTA, 0.05% bromophenol blue, and xylene cyanol. After denaturation at 85 °C for 5 min, 20 μL of the mixture was applied to a 12% polyacrylamide gel (39:1 acrylamide:bisacrylamide ratio) containing 1 × TBE buffer and 50 mL/L glycerol. The size of the gel was 10 cm × 9 cm × 0.1 cm (thickness). Electrophoresis was then performed for 4.5 h at 200 V using a buffer system containing 25 mM Tris and 192 mM glycine. The buffer temperature was maintained in a range of 16 °C-23 °C.
After electrophoresis, the gel was fixed for 30 min in 100 mL of a mixture consisting of 100 mL/L ethanol and 50 mL/L acetic acid, and then washed twice in distilled water. The gel was stained in 1 mL/L AgNO3 for 30 min, and then washed three times in distilled water. After washing, the gel was submerged in a mixture of 25 g/L Na2CO3 and 0.4 mL/L formalin, and then allowed to develop for 20 min-30 min until bands became visible. The reaction was stopped by transferring the stained gel into 100 mL/L acetic acid solution.
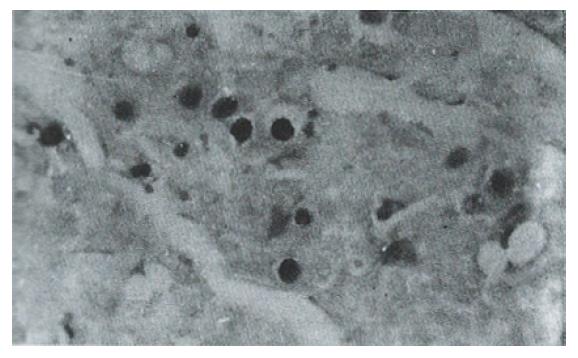
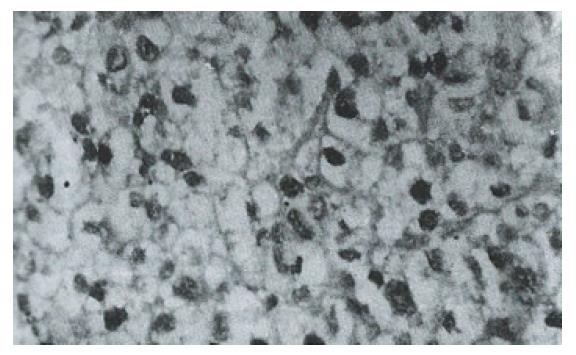
Sixteen of 38 HCC specimens were positive for P53 protein (42.1%), while the corresponding noncancerous tissues were all negative. The majority of malignant cells in all of the P53-positive specimens displayed a stained nucleus, and some also showed cytoplasmic staining. The distribution of p53-positive cells showed a single, focal or diffuse pattern (Figure 2 and Figure 3).
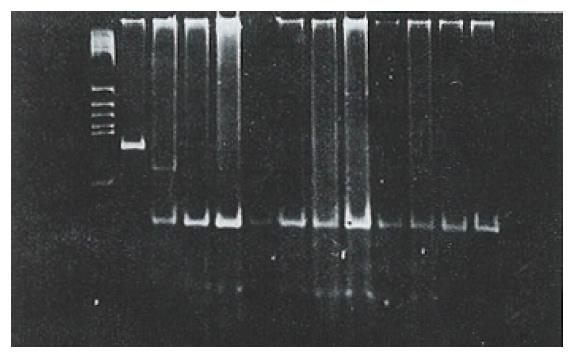
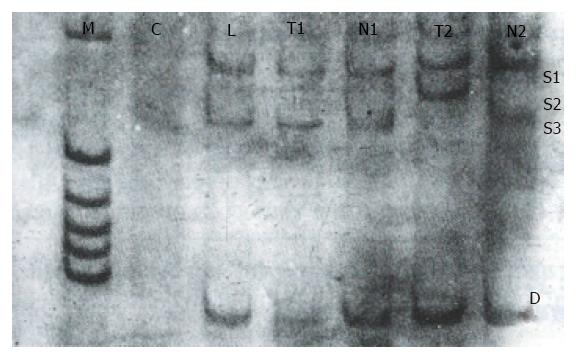
After digesting the PCR product with Hae III, seven (18.4%) of the 38 tissue samples were identified as mutant-types (136 bp and 27 bp fragments) (Figure 4). No mutations were found in 38 corresponding noncancerous tissue samples and 22 samples of normal lymphocytes. When the remaining 31 pairs of wild type HCC specimens were analyzed by PCR-SSCP-silver staining, two showed a mobility shift (Figure 5), indicating the presence of mutations in exon 7, rather than codon 249. Thus the mutation incidence in p53 exon 7 was 23.7% (9/38 samples). Among 9 mutant-type HCC specimens 8 specimens (88.9%) tested positive for P53 protein.
| Clinicopathological features | Number of cases | p53 expression(+) | p53 expression(-) | Significance |
| Tumor size (cm) | ||||
| < 3 | 3 | 1 | 2 | |
| 3-5 | 5 | 2 | 3 | a |
| > 5 | 30 | 13 | 17 | |
| Classification | ||||
| Trabecular | 15 | 5 | 10 | |
| Pseudoglandular | 16 | 8 | 8 | b |
| Compact | 7 | 3 | 4 | |
| Grade | ||||
| I | 3 | 1 | 2 | |
| II | 7 | 2 | 5 | c |
| III | 15 | 5 | 10 | |
| IV | 13 | 8 | 5 | |
| Metastasis | ||||
| Positive | 14 | 10 | 4 | |
| Negative | 24 | 6 | 8 | d |
Our results showed that overexpression of p53 was more frequent in cases of metastasized grade IV HCC. No significant relationship was found between p53 overexpression and either tumor size or histological classification.
| Clinicopathological features | Number ofcases | p53 expression (+) | p53 expression (-) | Significance |
| Tumor size (cm) | ||||
| < 5 | 8 | 0 | 8 | |
| > 5 | 30 | 9 | 21 | |
| Classification | ||||
| Trabecular | 15 | 4 | 11 | |
| Pseudoglandular | 16 | 3 | 13 | b |
| Compact | 7 | 2 | 7 | |
| Grade | ||||
| I | 3 | 0 | 3 | |
| II | 7 | 1 | 6 | c |
| III | 15 | 2 | 13 | |
| IV | 13 | 6 | 7 | |
| Metastasis | ||||
| Positive | 14 | 6 | 8 | |
| Negative | 24 | 6 | 21 | d |
In this study, p53 mutations were found more frequently in HCCs having a large size, a grade IV classification, and which had metastasized.
P53 protein has a very short half life in normal cells, and is thus undetectable by standard immunocytochemical methods. In contrast, p53 gene mutations in tumor cells usually result in the stabilization and accumulation of p53 protein in the nuclei, at levels which can be detected by immunohistochemistry techniques. Therefore, p53 overexpression detected by immunohistochemistry strongly suggests the presence of a p53 gene mutation[6]. Among the 9 mutant-type HCC specimens in this study, 8 specimens (88.9%) were positive for P53 protein, which confirms the above hypothesis. Because the location of a p53 point mutation can vary, DNA sequencing is essential to reveal this alteration of the gene. However, immunohistochemical techniques are more simple to perform, and especially useful when screening large numbers of specimens for the presence of mutations.
The presence of p53 mutations in HCC appears to vary with geographic location. In regions with a high risk for exposure to HBV and aflatoxins, (e.g., China[2] and South Africa[1]), p53 mutations have been reported in ~ 50% of HCC cases, and the majority of those mutations were found at codon 249. In Japan[7], where HBV infections are relatively common, but dietary aflatoxin intake is low, p53 mutations have been reported in 29% of HCC cases. In countries where neither HBV nor dietary aflatoxin are prevalent (e.g., France[8], Germany[9], and Great Britain[10]), mutated p53 is rarely found in HCCs, and there have been few or no mutations reported at codon 249 in the latter two countries[3,11]. While the results of molecular epidemiological studies have suggested that HBV may act synergistically with aflatoxin in the generation of p53 mutations in HCC, its mechanism of action remains obscure. In this study, we divided p53 mutations into two groups: (1) a tumor-specific mutation at codon 249, and (2) various scattered mutations at codons other than codon 249. Among 38 cases of HCC, 7 cases (18.4%) were found to have specific mutations by PCR-RFLP analysis. This 18.4% mutation rate was much lower than the 50% rate reported in Qidong[12], an area with a high exposure to aflatoxin, and much higher than the 2.2%rate reported in northern China[13], an area with low exposure to aflatoxin. Meanwhile, immunohistochemistry and PCR-SSCP analyses indentified 10 cases of HCC (26.3%) which showed scattered mutations. The overall frequency of p53 mutations in our geographic area was 44.7% (17/38 HCC cases), and these results are in agreement with the high prevalence of HBV and moderate exposure to aflatoxin in our area. We therefore conclude that HBV acts in conjunction with aflatoxin to generate p53 gene mutations in HCCs. Furthermore, dietary exposure to AFB1 may mainly contribute to the tumor-specific mutation, while HBV infection may account for other p53 gene mutations. As the most common tumor suppressor gene, p53 contributes to malignant transformation and the aggressiveness shown by several types of cancers. It was previously reported that p53 overexpression is correlated with the aggressiveness and metastasis of HCC[14]. Because P53 protein is not abnormally expressed in peri-cancerous liver tissue, the p53 gene probably becomes mutated after the hepatocellular carcinogenesis process has already been initiated. Both p53 overexpression and p53 mutations were more frequent in poorly-differentiated HCCs and cases of metastasized HCC, suggesting that the number of p53 alterations had increased as the HCC progressed. These findings indicate that p53 mutations represent one of the most important genetic changes in the development of HCC, and can serve as a molecular marker for high grade and metastatic HCC.
The authors thank Dr. Crawford L for his donation of monoclonal antibody Pab1801, and Dr. Lane DP for his donation of polyclonal antiserum CM-1.
This paper was presented at the 4th Congress, Asia Pacific Association of Societies of Pathologists, May 1995, Beijing.
Original title:
S- Editor: Ma JY L- Editor: Filipodia E- Editor: Li RF
| 1. | Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 872] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 2. | Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 969] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 3. | Unsal H, Yakicier C, Marçais C, Kew M, Volkmann M, Zentgraf H, Isselbacher KJ, Ozturk M. Genetic heterogeneity of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1994;91:822-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 109] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | van den Berg FM, Baas IO, Polak MM, Offerhaus GJ. Detection of p53 overexpression in routinely paraffin-embedded tissue of human carcinomas using a novel target unmasking fluid. Am J Pathol. 1993;142:381-385. [PubMed] |
| 5. | Wang D, Shi JQ. Antigen retrieval technique for immunohistochemical demonstration of p53 protein in paraffin sections. Disan Junyi Daxue Xuebao. 1994;16:52-54. |
| 6. | Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Engl J Med. 1993;329:1318-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 932] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 7. | Oda T, Tsuda H, Scarpa A, Sakamoto M, Hirohashi S. p53 gene mutation spectrum in hepatocellular carcinoma. Cancer Res. 1992;52:6358-6364. [PubMed] |
| 8. | Laurent-Puig P, Flejou JF, Fabre M, Bedossa P, Belghiti J, Gayral F, Franco D. Overexpression of p53: a rare event in a large series of white patients with hepatocellular carcinoma. Hepatology. 1992;16:1171-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Kress S, Jahn UR, Buchmann A, Bannasch P, Schwarz M. p53 Mutations in human hepatocellular carcinomas from Germany. Cancer Res. 1992;52:3220-3223. [PubMed] |
| 10. | Challen C, Lunec J, Warren W, Collier J, Bassendine MF. Analysis of the p53 tumor-suppressor gene in hepatocellular carcinomas from Britain. Hepatology. 1992;16:1362-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 60] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Wang D, Shi JQ. The progress of tumor suppressor gene in human hepatocellular carcinoma (Review). Foreign Medical Sciences. Genetics. 1994;17:240-244. |
| 12. | Scorsone KA, Zhou YZ, Butel JS, Slagle BL. p53 mutations cluster at codon 249 in hepatitis B virus-positive hepatocellular carcinomas from China. Cancer Res. 1992;52:1635-1638. [PubMed] |
| 13. | Zhu MH, Wang WL. Study of allotype and suppressor gene p53 exon 7 in hepatocellular carcinoma. Disi Junyi Daxue Xuebao. 1993;14:241-246. |
| 14. | Hsu HC, Tseng HJ, Lai PL, Lee PH, Peng SY. Expression of p53 gene in 184 unifocal hepatocellular carcinomas: association with tumor growth and invasiveness. Cancer Res. 1993;53:4691-4694. [PubMed] |