Published online Dec 14, 2013. doi: 10.3748/wjg.v19.i46.8764
Revised: September 11, 2013
Accepted: September 29, 2013
Published online: December 14, 2013
Processing time: 172 Days and 11.6 Hours
AIM: To evaluate the impact of Bmi-1 on cell senescence and metastasis of human gastric cancer cell line BGC823.
METHODS: Two pairs of complementary small hairpin RNA (shRNA) oligonucleotides targeting the Bmi-1 gene were designed, synthesized, annealed and cloned into the pRNAT-U6.2 vector. After DNA sequencing to verify the correct insertion of the shRNA sequences, the recombinant plasmids were transfected into BGC823 cells. The expression of Bmi-1 mRNA and protein was examined by reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting. The effects of Bmi-1 knockdown on cell senescence and metastasis were determined by the β-Gal activity assay and Boyden chamber assay, respectively.
RESULTS: The double-stranded oligonucleotide fragments of Bmi-1 short interfering RNA (siRNA) cloned into pRNAT-U6.2 vector conformed to the inserted sequence. RT-PCR and Western blotting indicated that the expression levels of Bmi-1 gene mRNA and protein were markedly decreased in transfected BGC823 cells with pRNAT-U6.2-si1104 and pRNAT-U6.2-si1356, especially in transfected BGC823 cells with pRNAT-U6.2-si1104, compared with two control groups (empty vector and blank group). In particular, Bmi-1 protein expression was almost completely abolished in cells transfected with the recombinant vector harboring shRNA targeting the sequence GGAGGAGGTGAATGATAAA (nt1104-1122). Compared with untransfected cells and cells transfected with the empty vector, the mean percentage of senescent cells increased and the number of cells passing through the Matrigel decreased in cells transfected with the recombinant vectors.
CONCLUSION: Silencing Bmi-1 by RNA interference can increase the senescent cell rate and effectively reduce the metastasis of gastric cancer cells.
Core tip: The overexpression of Bmi-1 contributes to the development of cancers. This study aimed at to evaluate the impact of Bmi-1 on the senescence and metastasis of human gastric cancer. The results demonstrated that inhibition of Bmi-1 gene expression can enhance the senescence of human gastric cancer cells and inhibit the invasion and metastasis of gastric cancer. This research has provided an indication that Bmi-1 inhibitors might be developed as new agents for gastric cancer.
- Citation: Gao FL, Li WS, Liu CL, Zhao GQ. Silencing Bmi-1 enhances the senescence and decreases the metastasis of human gastric cancer cells. World J Gastroenterol 2013; 19(46): 8764-8769
- URL: https://www.wjgnet.com/1007-9327/full/v19/i46/8764.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i46.8764
Bmi-1 (B lymphoma Mo-MLV insertion region 1 homolog), a member of the polycomb group (PcG), functions as a transcriptional repressor and presents with high expression in many tumors, indicating a poor prognosis[1,2]. Several lines of evidence suggest that Bmi-1 blocks cell senescence and proliferation[3,4], and the Bmi-1 gene is also associated with tumor invasion and metastasis[5]. Based on a list of genes on a wild-type and Bmi-1-deficient genetic background, Bmi-1 has been identified as a predictor of the response to therapy and survival in multiple types of cancer[6,7]. Therefore, this study intended to silence Bmi-1 in BGC823 cells by RNA interference, to observe the role of Bmi-1 in the senescence and metastasis of gastric cancer cells.
Short interfering RNA (siRNA) vector pRNAT-U6.2 was purchased from GenScript Inc. (Piscataway, NJ, United States), Bmi-1 antibody from Santa Cruz Biotechnology (CA, United States). BglII, HindIII and T4DNA ligase were obtained from Promega. BGC823 human gastric cancer cell lines were received from the Chinese Academy of Science. RPMI 1640 and fetal bovine serum were supplied by Gibco BRL (Grand Island, NY, United States). Liposomes LipofectAmineTM2000, G418, Trizol reagent and reverse transcription-polymerase chain reaction (RT-PCR) kit were purchased from Invitrogen (Carlsbad, CA, United States) and senescence β-galactosidase staining kit (Cell Signaling Technology, Beverly, MA, United States).
Selection of siRNA for Bmi-1 target sequence: The analysis and design of Promega siRNA target sequence scanned human Bmi-1 gene sequence (NM_005180) was based on the design principle of siRNA target sequence. The 19bp siRNA target sequences, including 1104nt-1122nt (GGAGGAGGTGAATGATAAA) and 1356nt-1374nt (GAGAGATGGACTGACAAAT), were selected as the target sequence after the BLAST homology analysis. Two oligonucleotide hairpin DNA single strands were synthesized (1104F and 1104R, 1356F and 1356R), adding BamHI and XhoI endonuclease residues at the two ends. Two oligonucleotide hairpin DNA single strands demonstrated the following:
1104F: 5’-GATCCGGAGGAGGTGAATGATAAATTCAAGAGATTTATCATTCACCTCCTCCTTTTTTC-3’,
1104R: 5’-TCGAGAAAAAAGGAGGAGGTGAATGATAAATCTCTTGAATTTATCATTCACCTCCTCCG-3’;
1356F: 5’-GATCCGAGAGATGGACTGACAAATTTCAAGA
GAATTTGTCAGTCCATCTCTCTTTTTTC-3’,
1356R: 5’-TCGAGAAAAAAGAGAGATGGACTGACAAATTCTCTTGAAATTTGTCAGTCCATCTCTCG-3’.
Reconstruction of siRNA vectors: The single-stranded DNA oligonucleotide (1104F and 1104R, 1356F and 1356R) was converted into a double-stranded DNA (si1104 and si1356) by conventional annealing, and reconnected overnight at 4 °C, utilizing 2 × reaction reconnected buffer (5 μL), linear pRNAT-U6.2 vector (1 μL), T4 ligase (1 μL) and annealing product (3 μL). The two recovered products were incubated at 16 °C for 16 h after addition of Solution I containing DNA ligase, and the resulting ligated products were used to transfect well-prepared competent E. coli DH5α. The whole transfection mix was plated onto a prewarmed LB-ampicillin (AMP) agar plate and then incubated at 37 °C for 12 h. Individual growing colonies were picked out and incubated at 37 °C for 12 h in LB broth containing AMP. Full length plasmid DNA was extracted from positive clones using a plasmid DNA extraction kit and then subject to testing for the presence of Bmi-1 with nuclease digestion using Bgl II, Hind III and T4DNA ligase.
Identification of recombinants: The recombinants were identified by PCR amplification, using primers PRNA-U6.2 FORWARD and PRNA-U6.2 REVERSE. PCR reaction was performed with 3 min of initial denaturation at 94 °C, 35 cycles of 45 s denaturation at 94 °C, 45 s annealing at 55 °C, 45 s extension at 72 °C, and finally 10 min extension at 68 °C. RT-PCR amplification products were electrophoresed and inspected on a 1.1% agarose gel, and recovered and purified by using DNA Gel recovery kit.
Transfection by liposome-mediated siRNA: The transfection process was according to the Lipofectamine™2000 instructions: a cell suspension containing 4-8 × 105 cells was added to 500 μL of growth medium with serum but without antibiotics; 0.8-1.2 μg DNA was added to 50 μL of medium without serum; 2 μL of Lipofectamine™ 2000 was added to 50 μL OptiMEM® I medium and incubated for 5 min at room temperature; the DNA-Lipofectamine™ 2000 complexes were added and incubated for 4 h at 37 °C in a CO2 incubator. Finally cells were assayed at 24-48 h post-transfection for the appropriate activity.
RT-PCR analysis: RT-PCR was carried out as described previously[8]. Cells were harvested and rinsed with phosphate-buffered saline (PBS) at corresponding time points and total RNA in the treated sections was extracted according to the total RNA extracting kit. A solution was added consisting of 10 mmol/L dNTP, 0.5 g/L oligo(dT), 40 U reverse transcriptase (m-mulv), 59 pH 8.3 RT buffer (250 mmol/L Tris-HCl, 250 mmol/L KCl, 20 mmol/L MgCl2, 50 mmol DTT) and deionized water. Total sample volume was 20 μL. Samples were incubated at 37 °C for 1 h and the reaction was stopped by heating at 70 °C for 10 min. Reverse transcriptase was used to synthesize the first-strand cDNA from an equal amount of the RNA sample following the manufacturer’s instructions. About 35-45 cycles of PCR reaction were used to cover the linear range of the PCR amplification. The Bmi-1 specific primers (forward 5’GGAGACCAGCAAGTATTGTCC 3’; reverse 5’GACCATTCCTTCTCCAGGTAT 3’) were used to amplify a 517 bp fragment of the Bmi-1 coding region. β-actin was used as an internal control to amplify a 268 bp fragment. The band densities were scanned with a densitometer (Bio-Rad, United States). The relative amount of mRNA in each sample was calculated from the densitometry ratio of Bmi-1 OD value/β-actin OD value.
Western blotting analysis: Western blotting was conducted according to the manufacturer’s instructions. The samples of each supernatant and the final pellets were heat-blocked for 5 min in a loading buffer (125 mmol/L Tris-HCl, 20%glycero1, 10%2-mercaptethanol, 4% SDS, 0.02% bromophenol blue, pH 6.8) and then subjected to electrophoresis on a 10%-20% Tris-glycine sulfate-polyacrylamide gel. The samples were then electronically transferred to a transfer membrane and blocked for 1 h in Tris-HCl buffered saline containing 5% skimmed milk and 0.1% Tween. Primary antibodies were incubated at 4-8 °C overnight in a TBS buffer containing 5% bovine albumin. The membrane was rinsed with TBS buffer containing 0.1% Tween 20, incubated with HRP-labeled second antibody for 2 h, and then stained with the detection reagents. Western blot analysis was performed as described previously to assess the protein expression level of Bmi-1 (1:200) and β-actin (1:100). Blots were developed with a SuperSignal ECL Western blotting Dura Substrate kit (Pierce Biotech, Rockford, IL, United States).
Senescence staining: Cell senescence β-galactosidase staining was carried out according to the manufacturer’s instructions. Growth medium was removed from the cells and the plate rinsed once with PBS (2 mL for a 35 mm well), followed by addition of 1mL of 1x Fixative Solution to each 35 mm well. Cells were allowed to fix for 10-15 min at room temperature. The plate was rinsed twice with PBS (2 mL for a 35 mm well). After addition of 1 mL of β-galactosidase staining solution to each 35 mm well, the plate was incubated at 37 °C overnight in a dry incubator. While the β-galactosidase staining solution was still on the plate, the cells were checked under a microscope (× 200 total magnification) for the development of blue color. Five visual areas were randomly selected and photographed to record the percentage of the senescent cells.
Cell migration and invasion assay: Serum-free 1640 medium containing Matrigel was added to the filter membrane of the upper chamber to prepare a gel at 37 °C for 2 h. The 200 μL supernatant of serum-free NIH3T3 cells was utilized as chemokines in the lower chamber. After adding 400 μL of cells (1 × 109/L) to the upper chamber, they were cultured at 37 °C for 24 h. Five visual areas in the lower chamber were randomly selected and the percentage of senescent cells was recorded with hematoxylin-eosin staining. Each group had five parallel experiments.
Western blotting and RT-PCR results were analyzed with scanning densitometry (Bio-Rad). Quantitative data were documented as the mean ± SD. The significance of the differences was analyzed using SPSS 13.0 software (SPSS Inc., Chicago, IL, United States), with significance at P < 0.05.
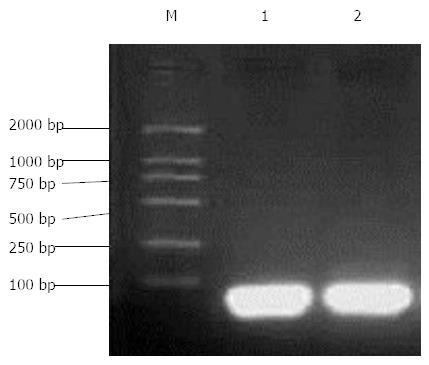
After annealing of hairpin single-stranded DNA for 1104 and 1356, the electrophoresis showed bright bands below 100 bp, consistent with the design (Figure 1).
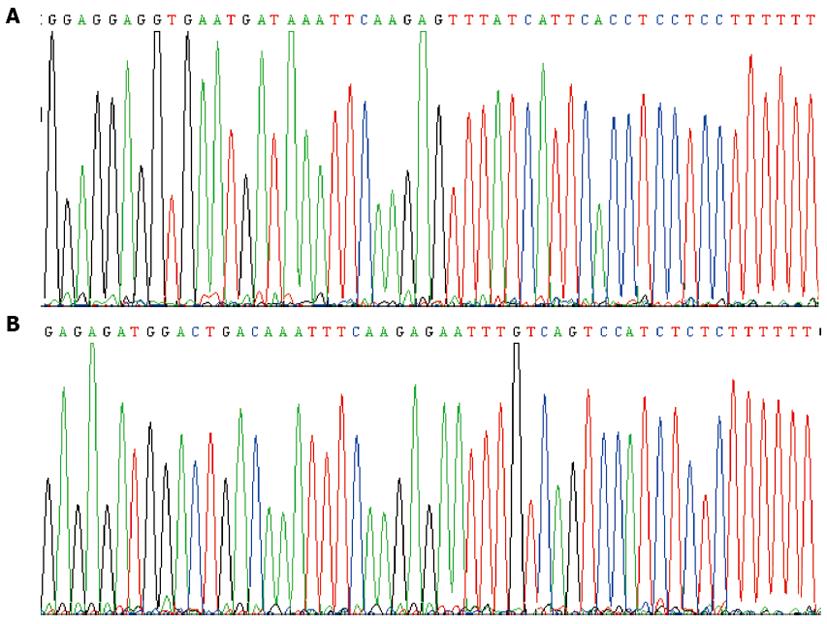
Two hairpin single-stranded DNA products (si1104 and si1356) were connected with pRNAT-U6.2 plasmid to transfect well-prepared competent E. coli DH5α. More than 10 transfected colonies grew on the Amp + LB culture plate. Ten transfected colonies were randomly selected. The DNA sequence of the inserted fragments was consistent with the designed positive recombinants (pRNAT-U6.2-si1104 and pRNAT-U6.2-si1356) (Figure 2).
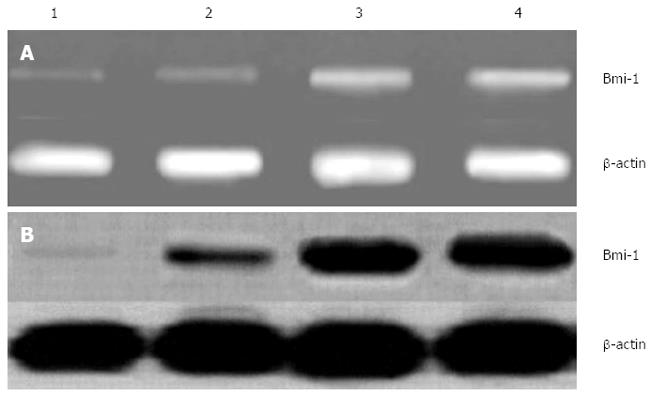
The expression of Bmi-1 mRNA was inhibited in transfected BGC823 cells with pRNAT-U6.2-si1104 and pRNAT-U6.2-si1356, especially in pRNAT-U6.2-si1104 transfected BGC823 cells, while two control groups (empty vector and blank groups) had significantly higher levels of Bmi-1 mRNA (P < 0.01) (Figure 3A).
There were high levels of Bmi-1 protein by Western blotting in non-transfected and transfected BGC823 cells with empty vector pRNAT-U6.2, compared with transfected BGC823 cells targeting Bmi-1 (pRNAT-U6.2-si1104 and pRNAT-U6.2 -si1356). while there was no Bmi-1 expression in the transfected BGC823 cells with pRNAT-U6.2-si1104 targeting Bmi-1 (P < 0.01) (Figure 3B).
The senescent rate of transfected BGC823 cells with pRNAT-U6.2-si1104 and pRNAT-U6.2-si1356 significantly increased compared with the non-transfected and transfected BGC823 cells with empty vector pRNAT-U6.2 (P < 0.01). The number of transfected BGC823 cells with pRNAT-U6.2-si1104 and pRNAT-U6.2-si1356 through the Matrigel significantly decreased, compared with the non-transfected and transfected BGC823 cells with empty vector pRNAT-U6.2 (P < 0.01) (Table 1).
This study aimed to investigate the impact of Bmi-1 on the senescence and metastasis of human gastric cancer cells, and our results indicate that inhibition of Bmi-1 gene expression can enhance the senescence of human gastric cancer cells and limit the invasion and metastasis of human gastric cancer cells.
Gastric cancer, the most common gastrointestinal malignancy, is the fourth most commonly diagnosed malignancy and the second leading cause of cancer-related death in the world[9]. Gastric cancer is often either asymptomatic or has nonspecific symptoms in its early stages. Once symptoms become apparent, the cancer has often reached an advanced stage and may also have metastasized and spread to other parts of the body. Accordingly, gastric cancer has a relatively poor prognosis since invasion and metastasis are important prognostic factors[10,11]. Currently, there is evidence that the incidence of gastric cancer is related to multiple oncogenes, such as C-myc, Ras, Hst and C-erbB-2[12-14]. The Bmi-1 gene, a polycomb gene (PcG), has been reported as an oncogene with high expression in cancers, and this may be related to high aggressiveness, such that overexpression of Bmi-1 is associated with poor prognosis[1,7]. Compelling research has supported that the expression of Bmi-1 decreases tumor cell senescence and proliferation, and increases tumor invasion and metastasis. The Bmi-1 gene can be synergistic with C-myc to induce cell metastasis and tumor formation[3,15,16]. This study demonstrated that the inhibition of Bmi-1 gene expression can increase the senescence of gastric cancer cells and slow down the invasion and metastasis of gastric cancer cells. It has provided further evidence of a role for Bmi-1 in the pathogenesis of gastric cancer.
The senescence β-galactosidase staining kit is designed to detect β-galactosidase activity at pH 6, a known characteristic of senescent cells not found in presenescent, quiescent or immortal cells[17,18]. Boyden chamber assays are used to measure cell invasion and various types of cell migration[19,20]. In this study, the incidence of senescent gastric cancer cells was most obvious when Bmi-1 expression was inhibited, according to β-galactosidase activity. Meanwhile, the number of gastric cancer cells through the Matrigel significantly decreased after inhibiting Bmi-1 expression in the Boyden chamber assay, indicating that the inhibition of Bmi-1 expression can limit the invasion and metastasis of gastric cancer cells. These results suggest that inhibition of Bmi-1 gene expression can enhance cell senescence and reduce the capability for cell invasion and metastasis.
In conclusion, we documented in the present study that silencing Bmi-1 by RNA interference enhances the senescent cell rate and effectively reduces the metastasis of gastric cancer cells. Many studies have shown that Bmi-1 is essential in multiple pathways in the pathogenesis of gastric cancer. Other reports have suggested that Bmi-1 inhibitors have therapeutic potential for gastric cancer through various mechanisms. The current has provided additional support for the notion that Bmi-1 inhibitors might be developed as new agents for gastric cancer.
Bmi-1 (B lymphoma Mo-MLV insertion region 1 homolog) has been reported as an oncogene that plays an important role in several types of cancer. The amplification and overexpression of Bmi-1 contribute to the development of many tumors and cancers, such as skin, prostate, breast, ovarian, and colorectal, as well as hematological malignancies. Whether Bmi-1 influences cell senescence and metastasis of human gastric cancer remains unknown. The aim of this study was to evaluate the impact of Bmi-1 on cell senescence and metastasis of the human gastric cancer cell line BGC823.
Bmi-1 is essential in multiple pathways in the pathogenesis of gastric cancer. The role of Bmi-1 on cell senescence and metastasis of human gastric cancer remains unclear.
The inhibition of Bmi-1 gene expression can enhance the senescence of gastric cancer cells and limit the invasion and metastasis of gastric cancer cells.
Bmi-1 inhibitors have therapeutic potential for gastric cancer through various mechanisms. This research has provided additional support for the notion that Bmi-1 inhibitors might be developed as new agents for gastric cancer.
This study demonstrated that the inhibition of Bmi-1 gene expression can increase gastric cancer cell senescence and inhibit invasive behavior in a well-accepted Boyden chamber model. The present study focused on the role of Bmi in cell senescence and metastasis. It would help to understand the mechanism of Bmi contribution to cancer progression. The data presented in this manuscript are quite good and very supportive of the hypothesis tested.
P- Reviewers: Giordano A, Higgins PJ, Takao S S- Editor: Zhai HH L- Editor: Cant MR E- Editor: Liu XM
| 1. | Tong YQ, Liu B, Zheng HY, He YJ, Gu J, Li F, Li Y. Overexpression of BMI-1 is associated with poor prognosis in cervical cancer. Asia Pac J Clin Oncol. 2012;8:e55-e62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Yin T, Wei H, Leng Z, Yang Z, Gou S, Wu H, Zhao G, Hu X, Wang C. Bmi-1 promotes the chemoresistance, invasion and tumorigenesis of pancreatic cancer cells. Chemotherapy. 2011;57:488-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Wang Y, Guan Y, Wang F, Huang A, Wang S, Zhang YA. Bmi-1 regulates self-renewal, proliferation and senescence of human fetal neural stem cells in vitro. Neurosci Lett. 2010;476:74-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164-168. [PubMed] |
| 5. | Guo BH, Feng Y, Zhang R, Xu LH, Li MZ, Kung HF, Song LB, Zeng MS. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer. 2011;10:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 178] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 6. | Xin T, Zhang FB, Sui GJ, Jin XM. Bmi-1 siRNA inhibited ovarian cancer cell line growth and decreased telomerase activity. Br J Biomed Sci. 2012;69:62-66. [PubMed] |
| 7. | Wang Y, Zhe H, Ding Z, Gao P, Zhang N, Li G. Cancer stem cell marker Bmi-1 expression is associated with basal-like phenotype and poor survival in breast cancer. World J Surg. 2012;36:1189-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Cai Z, Zhao Y, Yao S, Bin Zhao B. Increases in β-amyloid protein in the hippocampus caused by diabetic metabolic disorder are blocked by minocycline through inhibition of NF-κB pathway activation. Pharmacol Rep. 2011;63:381-391. [PubMed] |
| 9. | Al-Marzoqee FY, Khoder G, Al-Awadhi H, John R, Beg A, Vincze A, Branicki F, Karam SM. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int J Oncol. 2012;41:1733-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Kusano T, Shiraishi N, Shiroshita H, Etoh T, Inomata M, Kitano S. Poor prognosis of advanced gastric cancer with metastatic suprapancreatic lymph nodes. Ann Surg Oncol. 2013;20:2290-2295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Wang L, Wu Y, Lin L, Liu P, Huang H, Liao W, Zheng D, Zuo Q, Sun L, Huang N. Metastasis-associated in colon cancer-1 upregulation predicts a poor prognosis of gastric cancer, and promotes tumor cell proliferation and invasion. Int J Cancer. 2013;133:1419-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 12. | Han S, Kim HY, Park K, Cho HJ, Lee MS, Kim HJ, Kim YD. c-Myc expression is related with cell proliferation and associated with poor clinical outcome in human gastric cancer. J Korean Med Sci. 1999;14:526-530. [PubMed] |
| 13. | Qinyu L, Long C, Zhen-dong D, Min-min S, Wei-ze W, Wei-ping Y, Cheng-hong P. FOXO6 promotes gastric cancer cell tumorigenicity via upregulation of C-myc. FEBS Lett. 2013;587:2105-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Tahara E. [Oncogenes in human gastric carcinoma]. Gan To Kagaku Ryoho. 1989;16:2149-2155. [PubMed] |
| 15. | Piccinni E, Di Zenzo G, Maurelli R, Dellambra E, Teson M, Has C, Zambruno G, Castiglia D. Induction of senescence pathways in Kindler syndrome primary keratinocytes. Br J Dermatol. 2013;168:1019-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536-546. [PubMed] |
| 17. | Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187-195. [PubMed] |
| 18. | Itahana K, Itahana Y, Dimri GP. Colorimetric detection of senescence-associated β galactosidase. Methods Mol Biol. 2013;965:143-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 19. | Chen HC. Boyden chamber assay. Methods Mol Biol. 2005;294:15-22. [PubMed] |
| 20. | Kao WT, Lin CY, Lee LT, Lee PP, Hung CC, Lin YS, Chen SH, Ke FC, Hwang JJ, Lee MT. Investigation of MMP-2 and -9 in a highly invasive A431 tumor cell sub-line selected from a Boyden chamber assay. Anticancer Res. 2008;28:2109-2120. [PubMed] |