Published online Dec 14, 2013. doi: 10.3748/wjg.v19.i46.8659
Revised: July 26, 2013
Accepted: August 17, 2013
Published online: December 14, 2013
Processing time: 224 Days and 4.8 Hours
AIM: To investigated the molecular cause of very early-onset ulcerative colitis (UC) in an 18-mo-old affected child.
METHODS: We analysed the interleukin-10 (IL10) receptor genes at the DNA and RNA level in the proband and his relatives. Beta catenin and tumor necrosis factor-α (TNFα) receptors were analysed in the proteins extracted from peripheral blood cells of the proband, his relatives and familial adenomatous polyposis (FAP) and PTEN hamartoma tumor syndrome (PHTS) patients. Samples were also collected from the proband’s inflamed colorectal mucosa and compared to healthy and tumour mucosa collected from a FAP patient and patients affected by sporadic colorectal cancer (CRC). Finally, we examined mesalazine and azathioprine effects on primary fibroblasts stabilised from UC and FAP patients.
RESULTS: Our patient was a compound heterozygote for the IL10RB E47K polymorphism, inherited from his father, and for a novel point mutation within the IL10RA promoter (the -413G->T), inherited from his mother. Beta catenin and tumour necrosis factor α receptors-I (TNFRI) protein were both over-expressed in peripheral blood cells of the proband’s relatives more than the proband. However, TNFRII was over-expressed only in the proband. Finally, both TNFα-receptors were shown to be under-expressed in the inflamed colon mucosa and colorectal cancer tissue compared to healthy colon mucosa. Consistent with this observation, mesalazine and azathioprine induced, in primary fibroblasts, IL10RB and TNFRII over-expression and TNFRI and TNFα under-expression. We suggest that β-catenin and TNFRI protein expression in peripheral blood cells could represent molecular markers of sub-clinical disease in apparently healthy relatives of patients with early-onset UC.
CONCLUSION: A synergistic effect of several variant alleles of the IL10 receptor genes, inherited in a Mendelian manner, is involved in UC onset in this young child.
Core tip: We identified a novel point mutation within the interleukin-10 (IL10) receptor genes promoter (the -413G->T), associated with mRNA under-expression. We propose that this mutation has a synergistic effect with other variant alleles of IL10 receptor genes in very-early ulcerative colitis (UC) onset in this young child. β-catenin and tumour necrosis factor α receptors-I (TNFRI) protein were both over-expressed in peripheral blood cells of proband relatives, whereas TNFRII was over-expressed only in the proband. We suggest that β-catenin and TNFRI protein expression could represent molecular markers of sub-clinical disease in apparently healthy relatives of patients with early-onset UC.
- Citation: Galatola M, Miele E, Strisciuglio C, Paparo L, Rega D, Delrio P, Duraturo F, Martinelli M, Rossi GB, Staiano A, Izzo P, Rosa MD. Synergistic effect of interleukin-10-receptor variants in a case of early-onset ulcerative colitis. World J Gastroenterol 2013; 19(46): 8659-8670
- URL: https://www.wjgnet.com/1007-9327/full/v19/i46/8659.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i46.8659
Inflammatory bowel diseases (IBD) are chronic relapsing inflammatory disorders thought to result from an inappropriate and continuing inflammatory response to commensal microbes in a genetically susceptible host[1]. Crohn’s disease (CD) and ulcerative colitis (UC) are the two main clinicopathological subtypes of IBD, common in developed countries, affecting the quality of life of approximately 1.4 million individuals in the United States and 2.2 million people in Europe[2-4].
Accumulating data suggest that these disorders result from an inappropriate inflammatory response to intestinal microbes in a genetically susceptible host[5]. Active IBD is defined as an infiltration of the lamina propria by innate immune cells (neutrophils, macrophages, dendritic and natural killer T cells) and adaptive immune cells (B and T cells). Increased numbers and activation of these cells in the intestinal mucosa enhance local levels of tumour necrosis factor-α (TNFα) and several pro-inflammatory interleukins (IL)[5-8].
Genome-wide association studies (GWAS) have been successful in IBD, identifying 99 non-overlapping genetic risk loci, including 28 that are shared between CD and UC[9,10]. Analyses of the genes and genetic loci implicated in IBD show several pathways that are crucial for intestinal homeostasis, including barrier function, epithelial restitution, microbial defence, innate immune regulation, reactive oxygen species generation, autophagy, adaptive immunity regulation, endoplasmic reticulum stress and metabolic pathways associated with cellular homeostasis. Early studies have suggested the existence of both protective and predisposing alleles[11]. Again, many genetic changes might affect genetic regions other than coding regions, indicating that allele-specific gene-expression changes contribute to the disease risk[12].
The relative importance of each individual pathway in the pathogenesis of IBD has not been determined. There is enthusiasm for a model in which mucosal inflammation results from defective activity of Treg cells. In this model, effector T cells that react to the microbial flora or other GI antigens are kept in check by a population of regulatory cells; defects in these cells lead to GI inflammation. IL10 production by Treg cells appears to be required for suppression of colitis[13].
A recent study has demonstrated that IBD with an early onset can be monogenic. Mutations in IL10 or its receptor lead to a loss of IL10 function and cause severe intractable enterocolitis in infants and small children[14].
IL10R consists of two α (IL10RA) and two beta (IL10RB) molecules. IL10RA and IL10RB genes have been mapped on chromosomes 11q23.3 and 21q22, respectively, and many single-nucleotide polymorphisms (SNPs) have been identified[15]. Recently, Moran et al[16] identified IL10Rs polymorphisms that confer risk for developing very early-onset IBD. Each novel, nonsynonymous SNP was identified only in the heterozygous state, and none of the resulting amino acid changes were predicted to be deleterious by SIFT or Polyphen.
The aims of this work were to clarify the molecular basis of UC in an 18-mo-old affected child. To this aim, we investigated the pathogenetic mechanisms of IL10 pathway alteration in the onset of UC in the proband, and we clarified the molecular changes associated with them. Moreover, we propose β-catenin and tumour necrosis factor α receptors-I (TNFRI) as molecular bio-markers of subclinical disease among apparently healthy family members of the index case. Finally, we have investigated the effect of mesalazine and azathioprine, the main pharmacological therapy used for IBD treatment, on the expression of IL10 receptors, TNFα and TNFα receptors.
The proband, exhibiting UC, was referred by paediatric gastroenterologists to the laboratory for genetic analysis. He was admitted to the hospital for bloody diarrhoea, asthenia, fever and a severe anaemia (haemoglobin 3.7 g/dL). He underwent upper and lower GI endoscopy. The upper GI endoscopy did not reveal any macroscopic and/or microscopic sign of disease. Ileocolonoscopy showed a severe ulcerative pancolitis, (E4-S1) according to the Paris classification[17]. The colonoscopic grade of inflammation was characterised by the presence of marked erythema, absent vascular pattern, friability erosions, associated with spontaneous bleeding and ulcerations, suggesting a grade 3 according to the Mayo endoscopic score[18]. A severe grade of inflammation was confirmed histologically by the diffuse presence of a large number of neutrophilic leukocytes (> 50/HPF) with crypt abscesses and significant acute inflammation with ulcerations in lamina propria. The presence of granulomas was excluded at any colonic levels, as well as at level of the distal ileum.
The child was treated with blood transfusions, antibiotics and steroid therapy without improvement. A rescue therapy with cyclosporine followed by mesalazine and azathioprine was then started. His following clinical history was characterised by relapsing-remitting symptoms and by the lack of response to drugs. The proband’s mother referred episodes of bloody diarrhoea, but she refused colonoscopy.
Blood samples from proband and healthy family members were collected at the same hospital as the patient. Normal colorectal mucosa and colorectal cancer tissues were sampled from patients with FAP or sporadic colon cancer operated on the “Istituto Nazionale dei Tumori” in Naples.
Samples from all subjects who participated in the study were collected after being granted authorisation from the “Comitato etico per le attività Biomediche - Carlo Romano” of the University of Naples Federico II, with protocol number 120/10. Such authorisation is given only once the study has received ethical approval, and participants’ informed and written consent has been obtained.
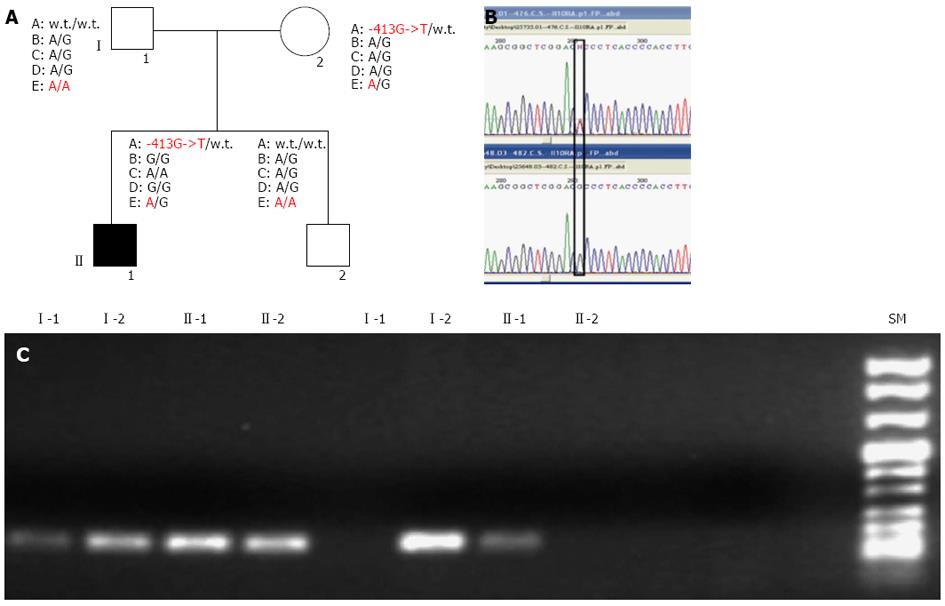
Reverse transcription polymerase chain reaction ofIL10RAandIL10RBof full length coding regions: Total RNA was extracted from 3 mL of peripheral blood cells of the UC patient and his healthy family members, using Trizol reagent (Invitrogen, Life Technologies, CA), cDNA was synthesised and 1 μL of the cDNA was amplified by reverse transcription polymerase chain reaction (RT-PCR) as previously described[19], using the following pairs of oligonucleotides: IL10RA-5’UTR-FP/IL10RA-3’UTR-RP; IL10RB-5’UTR-FP/IL10RB-3’UTR-RP. Two fragments of 2023 bp and 1197 bp, respectively, were produced. The PCR products were analysed on a 1% agarose gel in a tris-acetic acid (TAE)-EDTA standard buffer, and visualised by ethidium bromide staining (Table 1).
| RT-PCR of IL10RA and IL10RB of full length coding regions | ||
| IL10RA-5’UTR-FP: | GTCCCAGCCCAAGGGTAG | [NM_001558.3; start: + 5] |
| IL10RA-3’UTR-RP: | CACCCACATACCCTGCACTA | [NM_001558.3; start: + 2027] |
| IL10RB-5’UTR-FP: | GTCGTGTGCTTGGAGGAAG | [NM_000628.3; start: + 57] |
| IL10RB-3’UTR-RP: | GTGGCTAAGTCCAGGGTCTG | [NM_000628.3; start: + 1223] |
| Sequence analysis of IL10RA and IL10RB messenger/real time RT-PCR quantification analysis | ||
| IL10RA-5’UTRb-FP: | TCAGACGCTCATGGGACA | [NM_001558.3; start: + 132] |
| IL10RA-3’UTRb-RP: | CCCAGTGGACTTGCAGAAA | [NM_001558.3; start: + 1938] |
| IL10RA-3cFP: | AACTGGACCGTCACCAACAC | [NM_001558.3; start: + 405] |
| IL10RA-4cRP: | AATCTTCCCGAGGATGAAGC | [NM_001558.3; start: + 506] |
| IL10RA-6cFP: | AGCTACCCAGTGTCCTGCTC | [NM_001558.3; start: + 871] |
| IL10RA-7cRP: | CAAAAAGGCCTCCTCATCAA | [NM_001558.3; start: + 983] |
| IL10RB-5’UTRb-FP: | CATGGCGTGGAGCCTT | [NM_000628.3; start: + 99] |
| IL10RB-3’UTRb-RP: | GATGGTCTTGGCCCTTGTT | [NM_000628.3; start: + 1177] |
| IL10RB-4cFP: | GTGCAATACTGGAAAAACGGT | [NM_000628.3; start: + 565] |
| IL10RB-5cRP: | CCCTCGAACTTGAACACAATAA | [NM_000628.3; start: + 678] |
| Genomic PCR and sequencing | ||
| IL10RAp1-FP: | GCGGTTTGAGGCTCAGC | [NC_ 000011.9; start: + 117856447] |
| IL10RAp1-RP: | CAAGACGGAGGCTGAGGA | [NC_ 000011.9; start: + 117857234] |
| IL10RAp2-FP: | CTAGCAGGGGAAGAGCAGC | [NC_ 000011.9; start: + 117855574] |
| IL10RAp2-RP: | AACCTTCGTCTCCCAGGTTC | [NC_ 000011.9; start: + 117856355] |
| IL10RAp3-FP: | TGAGCCAAGTGACACAGAGG | [NC_ 000011.9; start: + 117855023] |
| IL10RAp3-RP: | TTGAACATATACCCTGCTGAAGAG | [NC_ 000011.9; start: + 117855810] |
| IL10RA-1FP: | CTGTCAGTCCCAGCCCAA | [NC_ 000011.9; start: + 17857104] |
| IL10RA-1RP: | TCTCCACTGGATGGAGAACTTTA | [NC_ 000011.9; start: + 117857327] |
| IL10RA-2FP: | TTGGTAAAATTGGGGTCATCA | [NC_ 000011.9; start: + 117859029] |
| IL10RA-2RP: | GCCCTCAGGCACTCACTTC | [NC_ 000011.9; start: + 117859328] |
| IL10RA-3FP: | AAGCTCGTTTCCAGTGCCTA | [NC_ 000011.9; start: + 117860120] |
| IL10RA-3RP: | GGCAGACATGGTGAGCTATG | [NC_ 000011.9; start: + 117860439] |
| IL10RA-4FP: | ACAAACCTGTGGCCAAGTTT | [NC_ 000011.9; start: + 117863822] |
| IL10RA-4RP: | CACACAAGGGTGCTTCCAG | [NC_ 000011.9; start: + 117864202] |
| IL10RA-5FP: | ATCACCTCTAAAGGCCCACC | [NC_ 000011.9; start: + 117864629] |
| IL10RA-5RP: | GGATGCAGAGCTATGTGAAGC | [NC_ 000011.9; start: + 117864993] |
| IL10RA-6FP: | TTTCATGGGACCAGAGTCCT | [NC_ 000011.9; start: + 117866223] |
| IL10RA-6RP: | CTGGCTGGGAGGAAAAGAG | [NC_ 000011.9; start: + 117864993] |
| IL10RA-7.1FP: | GCTCTCCTCCTGGGCCT | [NC_ 000011.9; start: + 117869338] |
| IL10RA-7.1RP: | CGGCCCTCAGAGTTTTGA | [NC_ 000011.9; start: + 117869854] |
| IL10RA-7.2FP: | ACCTGGGAGCAACAGGTG | [NC_ 000011.9; start: + 117869775] |
| IL10RA-7.2RP: | CGTGCCTAACTTCTGCCC | [NC_ 000011.9; start: + 117870445] |
| ARMS PCR of the -413G->T IL10-RA promoter mutation | ||
| IL10RA-ARMS-FP-N: | CCGGCACGCCAGGCAAAAGCGGCTCGGTCG | [NC_ 000011.9; start: + 117856738] |
| IL10RA-ARMS-FP-M: | CCGGCACGCCAGGCAAAAGCGGCTCGGTCT | [NC_ 000011.9; start: + 117856738] |
| IL10RA-ARMS-RP: | GCCTCCAGTGCCTTCGGATCAA | [NC_ 000011.9; start: + 117856897] |
| Gene copy number quantification of IL10RA gene | ||
| IL10RA-4cFP: | TCCTCGGGAAGATTCAGCTA | [NM_001558.3; start: + 493] |
| IL10RA-4c2RP: | TGCGAATGGCAATCTCATAC | [NM_001558.3; start: + 594] |
| IL10RA-7cFP: | ACTGAAGAGCCCCAGTTCCT | [NM_001558.3; start: + 1065] |
| IL10RA-7c2RP: | GCTGTCTGTGCTATTGCTGC | [NM_001558.3; start: + 1187] |
Sequence analysis of IL10RA and IL10RB mRNA: Sequence analysis of IL10RA and IL10RB full length coding regions was performed on amplified fragments from the cDNA of the proband and his healthy family members, using the following primer pairs, localised inside these regions: IL10RA-5’UTRb-FP; IL10RA-3’UTRb-RP; IL10RA-3cFP; IL10RA-4cRP; IL10RA-6cFP; IL10RA-7cRP; IL10RB-5’UTRb-FP; IL10RB-3’UTRb-RP; IL10RB-4cFP; IL10RB-5cRP (Table 1). The analysis was performed in a 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). For nucleotide numbering, the first A of the initiator ATG codon is nucleotide +1 of IL10RA and IL10RB mRNA sequences [GenBank Accession numbers: NM_001558.3 and NM_000628.3, respectively]; all oligonucleotides were obtained with primer-BLAST Software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
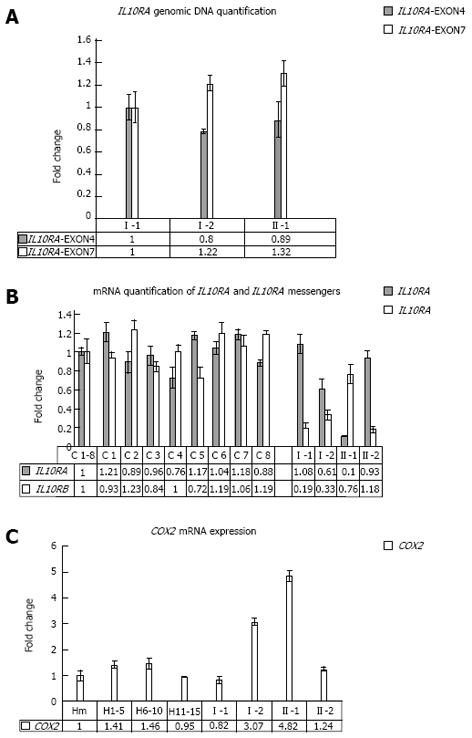
Real time RT-PCR quantification analysis: Real time PCR quantification analysis was performed for IL10RA and IL10RB messengers. The relative expression was calculated with the comparative Ct method. Patient numbering corresponds to that adopted in Figure 1A. Three millilitres of peripheral blood cells from the UC patient, his healthy family members and 8 healthy subjects were pelleted after erythrocyte lysis and resuspended in Trizol reagent. The mean value across all of the healthy samples (H1-8) was used as a calibrator to measure the relative expression. IL10RA and IL10RB mRNA quantification was carried out by amplifying fragments spanning the junctions between exons 3-4, for IL10RA messenger and exons 4-5 for IL10RB messenger, compared to the glucuronidase transcript fragment, using the oligonucleotides described above: IL10RA-3cFP/IL10RA-4cRP; IL10RB-4cFP/IL10RB-5cRP (Table 1). The quantitative real time assays were performed using the iCycler iQ Real Time Detection System BIO-RAD as previously described[19].
Genomic PCR and sequencing: Genomic DNA was extracted from 3 mL of peripheral blood cells of UC patient, using Nucleon BACC2 Kit (Amersham Biosciences). Genomic PCR and sequencing of all exons was performed for IL10RA gene, using oligonucleotides complementary to intronic neighbouring boundary regions of each exon, described in Table 1. The GenBank Accession number of IL10RA genomic sequence is: (NC_ 000011.9/gi:224589802). Mutational analysis of IL10RA promoter region, from bp -2159 to bp +1, was performed by PCR and sequencing. This region was amplified into three overlapping fragments of 788, 782 and 788 bp in molecular weight, respectively, using the following primer pairs: IL10RAp1-FP/IL10RAp1-RP; IL10RAp2-FP/IL10RAp2-RP; IL10RAp3-FP/IL10RAp3-RP (Table 1).
Amplification refractory mutation-PCR of the -413G->T IL10-RA promoter mutation: We set up an amplification refractory mutation-PCR (ARMS-PCR) reaction to analyse 200 DNA extracted from blood samples of control subjects apparently healthy, for the -413G->T promoter mutation identified in the UC proband and his mother. This ARMS reaction was performed with following oligonucleotide primers: IL10RA-ARMS-FP-N; IL10RA-ARMS-FP-M; IL10RA-ARMS-R (Table 1).
Gene copy number quantification of IL10RA gene: For the genomic quantification of IL10RA gene, specific amplified fragments were compared to a fragment of the exon 15 of MUTYH gene. For IL10RA specific quantification, two short fragments, one inside exon 4 and the other inside exon 7, were amplified, using the following primer pairs: IL10RA-4cFP/IL10RA-4c2RP; IL10RA-7cFP/IL10RA-7c2RP (Table 1). Patient numbering corresponds to that adopted in Figure 1A.
In silico analysis of the -413G->T point mutation was performed using the Patch 1.0 software. Patch is a pattern-based program for predicting transcription factor binding sites (TFBS) in DNA sequences. It uses the set of binding sites from TRANSFAC® Public 6.0 and is free online available at the web site: http://www.biobase-international.com/.
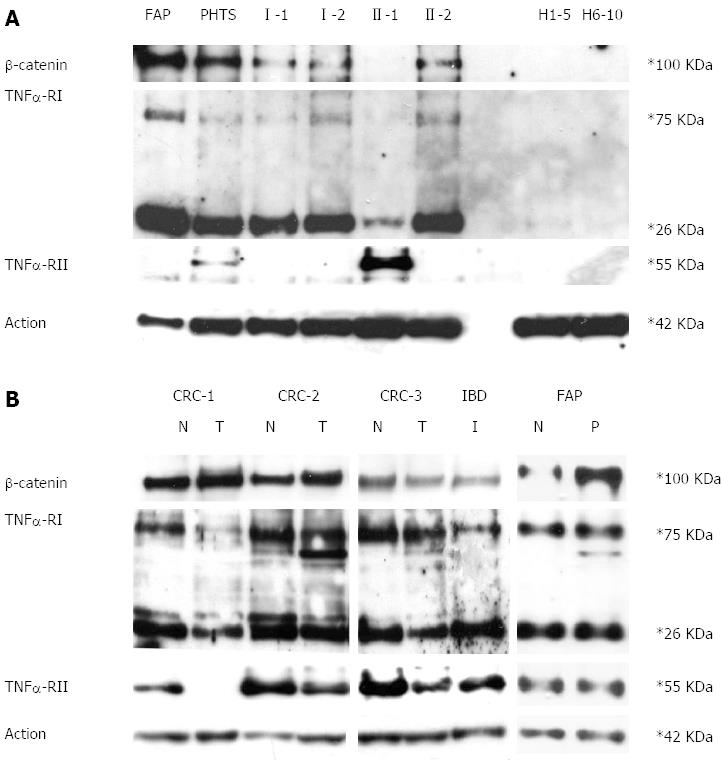
Western blotting assay ofβ-catenin, TNFRI and TNFRII proteins: Total protein was extracted from 3 mL of peripheral blood cells (approximately 5-7 × 103/mL cells) using Trizol reagent (Invitrogen, Life Technologies, CA) following the manufacturer’s instructions. Concentrations were determined and Western blotting assay was performed as previously described[19]. The primary antibody against amino-terminal β-catenin was from Cell Signaling Technology (Beverly, MA). Primary antibodies against TNFRI and TNFRII were from R&D System (R and D System, Minneapolis). The antibody against actin was from Santa Cruz (Santa Cruz, CA). H1-5 and H6-10 are mixes of healthy subjects. PHTS and FAP are two patients affected by PTEN hamartoma tumour syndrome and adenomatous polyposis coli syndrome, respectively. I-1, I-2, II-1 and II-2 are UC family members as reported in Figure 1A.
Real time PCR quantification analysis of COX2 mRNA: Real time PCR quantification analysis was performed for COX2 messengers. Relative expression was calculated with the comparative Ct method and normalised against the Ct of Glucuronidase (GUS) mRNA. The quantitative RNA real time assays were performed as described before. To better normalise the healthy values, we used three blood mixes as controls, each containing five samples collected from healthy subjects, for a total of fifteen controls. H1-5, H6-10, H11-15 are mixes of healthy subjects. Hm is the mean value among all healthy samples used as calibrator to measure the relative expression. Patient numbering corresponds to that adopted in Figure 1A.
Western blotting assay ofβ-catenin, TNFRI and TNFRII proteins: Total protein was extracted from the injured colorectal mucosa of the IBD proband and from healthy and tumour mucosa collected from patients affected by FAP and sporadic colorectal cancer using Trizol reagent (Invitrogen, Life Technologies, CA) following the manufacturer’s instructions. Western blotting analysis of β-catenin (amino-terminal antigen), TNFRI and TNFRII was performed as previously described.
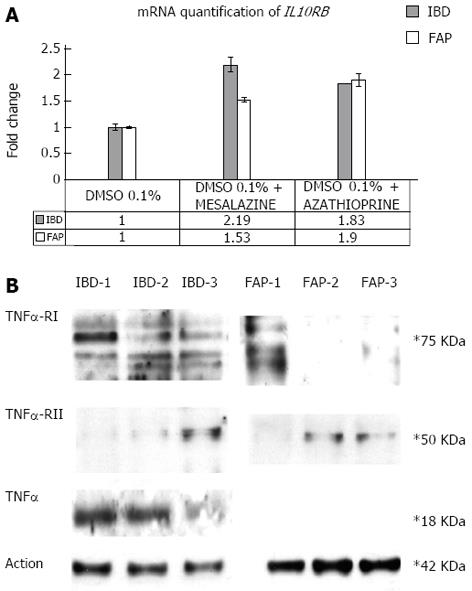
Incubation with mesalazine and azathioprine of established colon fibroblast culture: Samples of colorectal mucosa from IBD proband and one FAP patient were washed three times in PBS containing 300 U/mL penicillin, 300 μg/mL streptomycin, and 2.5 μg/mL amphotericin B (all from Gibco BRL, Karlsruhe, Germany), finely minced with scissors (tissue pieces of approximately 30 mm3) and digested in 2 mL 0.1% collagenase II (Boehringer Mannheim, Mannheim, Germany) in DMEM-15% FBS for 2 h at 37 °C, 5% CO2. The cell suspension was then collected by centrifugation, washed twice with serum-free DMEM medium, and subsequently cultured for 7 d in DMEM-15% FBS/CHANG C medium (1:1), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2.5 μg/mL amphotericin B (all from Gibco BRL, Karlsruhe, Germany). Primary fibroblasts from IBD and FAP patients were stabilised, cultured on plates, and incubated with mesalazine (30 mmol/L) and azathioprine (30 mmol/L) for 12 h, alternatively. A combination of real time PCR of IL10 receptors and Western blotting analysis of TNFα and TNFα receptors were performed as previously described.
Molecular screening of IL10RA and IL10RB, performed on the proband and his relatives, revealed the presence of multiple SNPs in the patient, inherited from his parents, as shown in Figure 1A.
Specifically, the proband was heterozygous for the IL10RB E47K polymorphism (rs2834167, A/G genotype), inherited from his father, described to be associated with a low level of specific mRNA expression (to the A allele). As shown in Figure 1, he was also carrier of an IL10RA promoter point mutation (the -413G->T point mutation), inherited from his mother and not previously described in literature. In silico analysis of this mutation, performed using the Patch 1.0 software, shows that it alters a binding site for the Sp1 transcription factor. This genomic variant represents a specific mutation of this IBD family because it was not identified in 200 healthy subjects. The proband’s father and his brother were both homozygous for IL10RB E47K polymorphism (rs rs.:2834167 A/A genotype; 47K/K), whereas his mother was heterozygous A/G. Only the proband and his mother were carriers of the -413G->T point mutation identified in the promoter region of the IL10RA gene. For the following SNPs of IL10RA, the rs2256111, localised in the exon 4 (c.549A->G; p.153Ala->Ala), the rs.:2229113, localised in the exon 7 (c.1051A->G; p.351Arg->Gly) and the rs.:9610, localised in the 3’UTR (c.2543G->A), the proband was homozygous G/G, G/G and A/A, respectively. These tree polymorphisms were A/G heterozygous in all other family members (Figure 1A). Using DNA real-time PCR for gene dosage of IL10RA gene, we ruled out the presence of intragenic or whole gene deletion (Figure 2A).
Associated with these genomic variants, we observed a under-expression of IL10RA and IL10RB mRNA in the proband compared to the average values of 8 healthy subjects, which segregates with each specific variant among the family members. In fact, as revealed by mRNA real-time quantification of both mRNAs of IL10 receptors shown in Figure 2B, only the proband and his mother, carriers of the -413G->T promoter point mutation, showed a decrease in IL10RA mRNA. In contrast, the proband’s father and his brother, both homozygous A/A for the IL10RB E47K polymorphism, show very low levels of IL10RB mRNA expression (fold change of approximately 0.19 and 0.18, respectively), whereas the proband and his mother, who were heterozygous A/G for this polymorphism, showed approximately 50% mRNA expression of the IL10RB compared to the mean value across eight healthy samples used as a calibrator (fold change of approximately 0.5 and 0.7 for the proband’s mother and the proband himself, respectively). Furthermore, only the proband and his mother showed COX2 overexpression, analysed in peripheral blood cells (Figure 2C).
As shown in Figure 3A, β-catenin and TNFRI protein were both over-expressed in the peripheral blood cells of the proband’s relatives more than the proband. In contrast, TNFRII was over-expressed only in the proband. None of these proteins were detectable in healthy controls. When investigated in colon mucosa, both TNFα receptors were observed to be under-expressed in the inflamed colon mucosa and colorectal cancer compared to healthy colon mucosa. In the FAP patient, normal colon mucosa and polyps express TNFα receptors at the same level. Furthermore, as expected, β-catenin expression is much higher in the polyp than in normal mucosa. (Figure 3B)
Finally, we show that after incubation with mesalazine and azathioprine of primary fibroblasts of the proband and of a FAP patient, drugs induce IL10RB mRNA and TNFRII protein over-expression, whereas TNFRI protein was under-expressed. A decrease of TNFα expression was also observed after incubation with azathioprine but not with mesalazine only in the IBD patient. Fibroblasts isolated from an FAP patient did not show any signal for TNFα hybridisation in our experimental conditions (Figure 4).
A recent study demonstrated that mutations in IL10 or its receptor lead to a loss of IL10 function and cause severe intractable enterocolitis in infants and small children[20,21]. In another approach to determining the genetic basis for these disorders, Moran et al[16] identified risk SNPs for very early onset IBD. Two SNPs, rs2228054 and rs2228055, were frequently found in the heterozygous state among IBD patients and inherited as a haplotype. The authors propose that the conferred risk may be due to one or both SNPs. Alternatively, the increased risk may reside in a regulatory region (e.g., promoter) in linkage disequilibrium with these SNPs and suggest that this risk haplotype exerts a mild phenotype in the general population resulting in disease only in the presence of other genetic variants or environmental triggers[16].
As suggested by Moran et al[16] and also described for other human diseases[22], our results confirm that early-onset IBD could be attributed to a synergistic effect of several variant alleles of the genes encoding IL10 receptors. These variants, alone, could only give rise to a sub-clinical manifestation of the disease. In fact, the proband’s father and his brother, both carriers of homozygous A/A polymorphism E47K for the IL10RB gene but without the -413G->T promoter mutation in the IL10RA gene, were apparently not affected. The proband’s mother shows a genotype very similar to the proband. In fact, they are both heterozygous for the E47K IL10RB gene polymorphism and for the -413G->T promoter mutation in the IL10RA gene. They show different mRNA expression for the IL10RA gene and quantitative real-time PCR revealed a 0.1 and 0.6-fold change for the IL10RA mRNA in the proband and his mother, respectively. This different gene expression could be due to other intragenic SNPs in the IL10RA gene whose alleles are different, such as, the rs.:2256111, localised in exon 4 (c.549A->G; p.153Ala->Ala), the rs.:2229113, localised in exon 7 (c.1051A->G; p.351Arg->Gly) and the rs.:9610, localised in the 3’UTR (c.2543G->A), that were homozygous G/G, G/G and A/A in the proband but A/G heterozygous in all other family members. However, we cannot rule out other gene expression regulatory mechanisms. Possibly due to the different IL10RA mRNA expression, the proband’s mother has not developed the disease. However, she referred to an episode of rectal bleeding and shows increased levels of COX2 mRNA expression in peripheral blood cells.
In a recent study, 66 early onset IBD patients were analysed. The authors identified 16 patients with loss-of-function mutations in the IL10 or IL10R genes. A variety of mutations were discovered. Most patients were born from consanguineous parents and they carried homozygous biallelic mutations (point mutations or deletions). However, some patients also presented compound heterozygous mutations. Genotype/phenotype correlations were not clearly observed. In fact, siblings sharing the same homozygous IL10RB mutation showed a remarkably distinct level of disease severity, suggesting that the phenotypic manifestation is dependent on other intrinsic or extrinsic factors that remain presently unknown[21,23].
Non-coding single nucleotide polymorphisms (SNPs) can be associated with qualitative and quantitative changes. Furthermore, genetic changes may affect transcription-factor-binding sequences, locus accessibility, translational efficiency and trans-regulators such as noncoding RNAs and microRNAs[12]. Cis- or trans-expression quantitative trait loci are detected for approximately half of the IBD risk regions, indicating that allele-specific gene-expression changes contribute to disease risk[24].
Unexpectedly, we observed β-catenin and TNFRI protein over-expression in the peripheral blood cells of the proband’s apparently healthy relatives more than in the proband himself. FAP and PHTS patients, but not healthy subjects, also expressed this protein, as previously described[19]. Therefore, we suggest that these proteins could represent a good candidate for molecular markers of sub-clinical disease in relatives of patients with UC. Previous studies showed that faecal calprotectin concentration in patients with CD and relatives differed significantly from controls, suggesting that there is a high prevalence of subclinical disease in first-degree relatives of these patients. This result conforms to an additive inheritance pattern in which the genetic basis for this abnormality may represent a risk factor for CD and UC[25,26].
Because no therapeutic approach was successful in patients who are carriers of IL10 pathway alterations, we investigated the effect of mesalazine and azathioprine on the expression of IL10 receptors, TNFα and TNFα receptors. In agreement with our hypothesis, we found TNFRI under-expression and TNFRII and IL10RB over-expression in primary fibroblasts incubated with mesalazine and azathioprine, in both the UC and FAP patients. In the UC patient only, azathioprine, but not mesalazine, induces a TNFα decrease.
These observations could suggest that these drugs are only able to partially restore IL10 pathway function in UC, by activation of IL10RB, but not IL10RA, transcription. On the other hand, under-expression of TNFRI and over-expression of TNFRII could increase the risk of colorectal cancer-associated colitis in UC patients. As described by Chang et al[27], TNFRI has tumour suppressor activity in the context of colitis-associated cancer, and the role of TNFRII in cell proliferation is well known.
Current therapeutic strategies for paediatric IBD include the use of exclusive enteral nutrition, corticosteroids, mesalamine, sulfasalazine, immunomodulators (azathioprine, 6-mercaptopurine, methotrexate) and anti-TNFα-antibodies[22,28]. Aminosalicylates are the undisputed first-line option for treating and maintaining remission in UC[29]. However, the role that these drugs may play in the management of Crohn’s disease has been controversial. Thiopurine drugs, azathioprine and mercaptopurine, have been shown to be effective in inducing and maintaining remission in IBD[30]. Most epidemiological studies have shown that the chronic use of 5-ASA in IBD has chemopreventive effects on the development of CRC[14,31], although some studies failed to show this, as described by Velayos et al[32].
TNF signals via two cell surface receptors, TNFRI and TNFRII, resulted in several, sometimes opposing, cellular responses that vary by context and cell nature[33,34]. In the colonic mucosa, TNF is involved in both cell survival and cell death[35]. Additionally, increased levels of TNF have been found in the setting of cancers, including those of the pancreas, skin, and ovaries[36]. With specific regard to colon carcinogenesis, TNF activity has been shown both to promote and to protect from neoplastic transformation[37-39] and there are case studies of development of cancer in other organ systems (lymphatic and skin) following the use of anti-TNF for IBD or rheumatological disease[40]. For this reason, we investigated protein expression of TNF receptors in colon mucosa of the UC patient compared to that of normal and cancer colon mucosa from patients affected by FAP and sporadic colorectal cancer. In agreement with the hypothesis suggested by Chang et al[27] about the tumour suppressor activity of TNFRI in the context of colitis-associated carcinogenesis, we found not only a decrease in the expression of TNFRI but also of TNFRII in colorectal cancer when compared to normal colon mucosa for each patient. The expression of TNF receptor proteins in colon mucosa of our UC patient was at an intermediate level between that observed in colorectal tumour tissue and normal mucosa of CRC patients.
In conclusion, our results, in agreement with data from recently published literature[5,16,22], indicate that early-onset UC could be caused by a synergistic effect of more variant alleles of the IL10 receptors gene, resulting in alteration of the IL10 pathway. In our opinion, a dosage model of nonallelic non-complementation fits well with this case, whereby mutations in two different genes can behave as alleles of the same locus by causing or exacerbating the same phenotype. However, we cannot exclude, as described for others syndromes, that different mechanisms, such as alternative splicing mechanisms[41,42] or allelic variants of modifier genes, could contribute to the observed phenotypic variability[22].
In addition, we suggest that the expression of β-catenin and TNFRI protein could represent molecular markers of sub-clinical disease in apparently healthy relatives of patients. Recent findings suggest that chronic inflammation in IL10-/- mice increased P-β-catenin552 expression. Moreover, TNFRI exerts its tumour suppressor activity by modulating activation of β-catenin and controlling epithelial proliferation[43]. It clearly appears that classical therapeutic approaches do not seem adequate for IBD patients who are carriers of IL10 pathway alterations because under-expression of TNFRI signalling would confer increased risk of developing colitis associated-carcinoma. Allogenic hematopoietic stem cell transplantation could represent a causal therapeutic approach for IL10R-deficient patients, useful for the treatment of the intractable ulcerating enterocolitis of the infant, as recently suggested[14,15,20-22].
Inflammatory bowel diseases (IBD) are chronic relapsing inflammatory disorders thought to result from an inappropriate and continuing inflammatory response to commensal microbes in a genetically susceptible host. Mutations in interleukin-10 (IL10) or its receptor lead to a loss of IL10 function and cause severe intractable enterocolitis in infants and small children.
Increased numbers and activation of immune cells in the intestinal mucosa enhance local levels of tumour necrosis factor-α (TNFα) and several proinflammatory IL. Recent work has demonstrated that IBD with an early onset can be monogenic and IL10 polymorphisms have been associated with IBD in genome-wide association studies. The aims of this work were to clarify the molecular basis of disease in this young child, shedding light on a synergistic effect of IL10RA and IL10RB polymorphisms. The authors also assessed the possible presence and inheritance of subclinical intestinal inflammation in apparently healthy relatives of this patient with ulcerative colitis (UC).
Recent studies have shown that loss-of-function mutations in IL10RA, IL10RB and IL10 genes, in immunodeficient patients, are associated with severe, infantile-onset IBD. In particular, literature reports have highlighted the role of IL10RA polymorphisms in the risk for developing very early onset UC. This is the first study reporting that IL10RA polymorphisms could have synergistic effect with those of IL10RB. The authors propose that these risk polymorphisms exert a mild phenotype in the general population resulting in disease only in the presence of other genetic variants in the IL10RA or IL10RB. Furthermore, these observations would suggest an inherited abnormality of beta catenin and TNFRI in the proband’s relatives.
This work expands the understanding of the complex inheritance pattern of very early onset ulcerative colitis. It seems possible that the subclinical phenotypic manifestations identified in the first-degree relatives of the proband represents the consequence of inherited defects of IL10R genes, which then represent one of the risk factors for the disease. This study could contribute to identifying at-risk families for very early onset UC allowing clinicians to perform genetic tests and appropriate care.
IL10 is an anti-inflammatory cytokine secreted by a variety of cell types and is critical for maintaining immune homeostasis in the gastrointestinal tract. IL10 activates downstream signalling by binding to IL10R, comprised of two α subunits (encoded by IL10RA) and two beta subunits (encoded by IL10RB).
The authors investigated the molecular cause of very early-onset inflammatory bowel disease in an 18-mo-old child as well as his relatives. They concluded that a synergistic effect of several variant alleles of the IL10 receptor genes, inherited in a Mendelian manner, is involved in IBD onset in this young child. This study supports a special enthusiasm about the potential power of genomics to define the aetiology and/or phenotype of diseases. When a single specific case or family is studied, the discovery of new functional polymorphisms and the functional consequences of these mutations deserves attention even if the functional characterisation and the real pathogenic contribution of susceptible genes are hard to assess in complex disorders such as IBD.
P- Reviewers: Corleto VD, Yamakawa M S- Editor: Zhai HH L- Editor: A E- Editor: Liu XM
| 1. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1882] [Article Influence: 134.4] [Reference Citation Analysis (2)] |
| 2. | Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729-1737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 406] [Article Influence: 29.0] [Reference Citation Analysis (1)] |
| 3. | Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self Nonself. 2010;1:299-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 157] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 4. | Bouguen G, Chevaux JB, Peyrin-Biroulet L. Recent advances in cytokines: therapeutic implications for inflammatory bowel diseases. World J Gastroenterol. 2011;17:547-556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1967] [Cited by in RCA: 2202] [Article Influence: 137.6] [Reference Citation Analysis (6)] |
| 6. | Bamias G, Nyce MR, De La Rue SA, Cominelli F. New concepts in the pathophysiology of inflammatory bowel disease. Ann Intern Med. 2005;143:895-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Andoh A, Yagi Y, Shioya M, Nishida A, Tsujikawa T, Fujiyama Y. Mucosal cytokine network in inflammatory bowel disease. World J Gastroenterol. 2008;14:5154-5161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 85] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 8. | Fantini MC, Monteleone G, Macdonald TT. New players in the cytokine orchestra of inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1419-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2110] [Cited by in RCA: 2006] [Article Influence: 133.7] [Reference Citation Analysis (0)] |
| 10. | Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1095] [Cited by in RCA: 1048] [Article Influence: 74.9] [Reference Citation Analysis (1)] |
| 11. | Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, Amininejad L, Cleynen I, Colombel JF, de Rijk P, Dewit O. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 12. | Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667-11672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2352] [Article Influence: 147.0] [Reference Citation Analysis (0)] |
| 13. | MacDonald TT, Monteleone I, Fantini MC, Monteleone G. Regulation of homeostasis and inflammation in the intestine. Gastroenterology. 2011;140:1768-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 206] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 14. | Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1149] [Cited by in RCA: 1092] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 15. | Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894-5903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 310] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 16. | Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, Wolters VM, Bandsma RH, Mouzaki M, Zachos M. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 189] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 17. | Levine A, Griffiths A, Markowitz J, Wilson DC, Turner D, Russell RK, Fell J, Ruemmele FM, Walters T, Sherlock M. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1216] [Cited by in RCA: 1144] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 18. | Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1958] [Cited by in RCA: 2250] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 19. | Galatola M, Paparo L, Duraturo F, Turano M, Rossi GB, Izzo P, De Rosa M. Beta catenin and cytokine pathway dysregulation in patients with manifestations of the “PTEN hamartoma tumor syndrome”. BMC Med Genet. 2012;13:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Glocker EO, Kotlarz D, Klein C, Shah N, Grimbacher B. IL-10 and IL-10 receptor defects in humans. Ann N Y Acad Sci. 2011;1246:102-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 21. | Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, Pfeifer D, Kreipe H, Pfister ED, Baumann U. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 346] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 22. | Duraturo F, Liccardo R, Cavallo A, De Rosa M, Grosso M, Izzo P. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: probability of synergistic effects. Int J Cancer. 2011;129:1643-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, Li H. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 188] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 25. | Thjodleifsson B, Sigthorsson G, Cariglia N, Reynisdottir I, Gudbjartsson DF, Kristjansson K, Meddings JB, Gudnason V, Wandall JH, Andersen LP. Subclinical intestinal inflammation: an inherited abnormality in Crohn’s disease relatives? Gastroenterology. 2003;124:1728-1737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 111] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Montalto M, Curigliano V, Santoro L, Armuzzi A, Cammarota G, Covino M, Mentella MC, Ancarani F, Manna R, Gasbarrini A. Fecal calprotectin in first-degree relatives of patients with ulcerative colitis. Am J Gastroenterol. 2007;102:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Chang F, Lacey MR, Bouljihad M, Höner Zu Bentrup K, Fortgang IS. Tumor necrosis factor receptor 1 functions as a tumor suppressor. Am J Physiol Gastrointest Liver Physiol. 2012;302:G195-G206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Van Assche G, Dignass A, Reinisch W, van der Woude CJ, Sturm A, De Vos M, Guslandi M, Oldenburg B, Dotan I, Marteau P. The second European evidence-based Consensus on the diagnosis and management of Crohn’s disease: Special situations. J Crohns Colitis. 2010;4:63-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 547] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 29. | Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults: American College Of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol. 2010;105:501-523; quiz 524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 899] [Cited by in RCA: 942] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 30. | Gisbert JP, Chaparro M, Gomollón F. Common misconceptions about 5-aminosalicylates and thiopurines in inflammatory bowel disease. World J Gastroenterol. 2011;17:3467-3478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Koelink PJ, Robanus-Maandag EC, Devilee P, Hommes DW, Lamers CB, Verspaget HW. 5-Aminosalicylic acid inhibits colitis-associated but not sporadic colorectal neoplasia in a novel conditional Apc mouse model. Carcinogenesis. 2009;30:1217-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 32. | Velayos FS, Terdiman JP, Walsh JM. Effect of 5-aminosalicylate use on colorectal cancer and dysplasia risk: a systematic review and metaanalysis of observational studies. Am J Gastroenterol. 2005;100:1345-1353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 379] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 33. | Ebach DR, Newberry R, Stenson WF. Differential role of tumor necrosis factor receptors in TNBS colitis. Inflamm Bowel Dis. 2005;11:533-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 255] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 34. | Ebach DR, Riehl TE, Stenson WF. Opposing effects of tumor necrosis factor receptor 1 and 2 in sepsis due to cecal ligation and puncture. Shock. 2005;23:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Corredor J, Yan F, Shen CC, Tong W, John SK, Wilson G, Whitehead R, Polk DB. Tumor necrosis factor regulates intestinal epithelial cell migration by receptor-dependent mechanisms. Am J Physiol Cell Physiol. 2003;284:C953-C961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Pezzilli R, Corsi MM, Barassi A, Morselli-Labate AM, Dogliotti G, Casadei R, Corinaldesi R, D’Eril GM. The role of inflammation in patients with intraductal mucinous neoplasm of the pancreas and in those with pancreatic adenocarcinoma. Anticancer Res. 2010;30:3801-3805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 759] [Reference Citation Analysis (0)] |
| 37. | Balkwill F. Tumor necrosis factor or tumor promoting factor? Cytokine Growth Factor Rev. 2002;13:135-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 350] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 38. | Grimm M, Lazariotou M, Kircher S, Höfelmayr A, Germer CT, von Rahden BH, Waaga-Gasser AM, Gasser M. Tumor necrosis factor-α is associated with positive lymph node status in patients with recurrence of colorectal cancer-indications for anti-TNF-α agents in cancer treatment. Cell Oncol (Dordr). 2011;34:315-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Popivanova BK, Kitamura K, Wu Y, Kondo T, Kagaya T, Kaneko S, Oshima M, Fujii C, Mukaida N. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118:560-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 456] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 40. | Ochenrider MG, Patterson DJ, Aboulafia DM. Hepatosplenic T-cell lymphoma in a young man with Crohn‘s disease: case report and literature review. Clin Lymphoma Myeloma Leuk. 2010;10:144-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 41. | De Rosa M, Galatola M, Borriello S, Duraturo F, Masone S, Izzo P. Implication of adenomatous polyposis coli and MUTYH mutations in familial colorectal polyposis. Dis Colon Rectum. 2009;52:268-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 42. | De Rosa M, Morelli G, Cesaro E, Duraturo F, Turano M, Rossi GB, Delrio P, Izzo P. Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript. Gene. 2007;395:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Lee G, Goretsky T, Managlia E, Dirisina R, Singh AP, Brown JB, May R, Yang GY, Ragheb JW, Evers BM. Phosphoinositide 3-kinase signaling mediates beta-catenin activation in intestinal epithelial stem and progenitor cells in colitis. Gastroenterology. 2010;139:869-881, 881.e1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |