Published online Nov 21, 2013. doi: 10.3748/wjg.v19.i43.7758
Revised: September 20, 2013
Accepted: September 29, 2013
Published online: November 21, 2013
Processing time: 201 Days and 12.5 Hours
AIM: To investigate the function of microRNA-143 (miR-143) in gastric cancer and explore the target genes of miR-143.
METHODS: A quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis was performed to evaluate miR-143 expression in gastric cancer cell lines. After transfecting gastric cancer cells with miR-143-5p and miR-143-3p precursors, Alamar blue and apoptosis assays were used to measure the respective proliferation and apoptosis rates. Cyclooxygenase-2 (COX-2) expression was determined by real-time RT-PCR and Western blot assays after miR-143 transfection. Reporter plasmids were constructed, and a luciferase reporter assay was used to identify the miR-143 binding site on COX-2.
RESULTS: Both miR-143-5p and miR-143-3p were significantly downregulated in multiple gastric cancer cell lines. Forced miR-143-5p and miR-143-3p expression in gastric cancer cells produced a profound cytotoxic effect. MiR-145-5p transfection into gastric cancer cells resulted in a greater growth inhibitory effect (61.23% ± 3.16% vs 46.58% ± 4.28%, P < 0.05 in the MKN-1 cell line) and a higher apoptosis rate (28.74% ± 1.93% vs 22.13% ± 3.31%, P < 0.05 in the MKN-1 cell line) than miR-143-3p transfection. Further analysis indicated that COX-2 expression was potently suppressed by miR-143-5p but not by miR-143-3p. The activity of a luciferase reporter construct that contained the 3’-untranslated region (UTR) of COX-2 was downregulated by miR-143-5p (43.6% ± 4.86%, P < 0.01) but not by miR-143-3p. A mutation in the miR-145-5p binding site completely ablated the regulatory effect on luciferase activity, which suggests that there is a direct miR-145-5p binding site in the 3’-UTR of COX-2.
CONCLUSION: Both miR-143-5p and miR-143-3p function as anti-oncomirs in gastric cancer. However, miR-143-5p alone directly targets COX-2, and it exhibits a stronger tumor suppressive effect than miR-143-3p.
Core tip: MicroRNA-143 (miR-143) has been reported to be a tumor suppressor. However, the functions of miR-143-5p and miR-143-3p have never been compared. In this study, we found that both miR-143-5p and miR-143-3p function as tumor suppressors in gastric cancer; however, miR-143-5p alone directly targets cyclooxygenase-2, and it exhibits a stronger tumor suppressive effect than miR-143-3p.
- Citation: Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol 2013; 19(43): 7758-7765
- URL: https://www.wjgnet.com/1007-9327/full/v19/i43/7758.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i43.7758
MicroRNAs (miRNAs) are short (19-24 nt) non-coding RNAs that control target mRNA translation and stability by binding to regulatory sites that are mostly located in the 3′-untranslated region (UTR) of transcripts[1]. Numerous miRNAs have been shown to display tumor suppressor activity, while others reportedly act as oncogenes[2]. The expression levels of these RNAs are altered in many human tumors, resulting in distinct miRNA networks in various tumor types[3]. Some targets of these miRNAs have been identified, but many of the critical cancer proteins and pathways that they regulate remain unknown.
MiR-143 is considered a pivotal regulator of gene expression because it directly targets multiple mRNAs that code proteins involved in cell proliferation, differentiation, survival and apoptosis, including cyclooxygenase-2 (COX-2)[4], V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS)[5,6], B-cell lymphoma 2 (Bcl-2)[7,8], plasminogen activator inhibitor-1 (PAI-1)[9], myosin VI (MYO6)[10], matrix metallopeptidase 13 (MMP-13)[11], DNA (cytosine-5)-methyltransferase 3A (DNMT3A)[12] and E twenty-six-like transcription factor 1 (ELK1)[13]. The relevance of miR-143 as a putative cancer biomarker is increasing, because this miRNA is downregulated in various human tumors and can suppress tumor growth in cancers of the urogenital system[7,9,10], digestive system[14-16], respiratory system[17,18] and nervous system[19], as well as some sarcomas[11,20] and B-cell malignancies[21]. MiR-143 expression has also been reported to be downregulated in human gastric cancer tissues and cell lines[22]. The expression of miR-143 is significantly decreased in stage IV gastric cancer, compared to stages I and II cancers[23]. However, the role of miR-143 in gastric cancer and the underlying mechanisms require further investigation.
Among the target genes regulated by miR-143, COX-2 is particularly important. COX, also known as prostaglandin (PG) H2 synthase, is the rate-limiting enzyme in the conversion of arachidonic acid into PGs. COX-2 expression in cells and animal models is associated with tumor cell growth and metastasis, enhanced cellular adhesion and apoptosis inhibition[24]. Pharmacologic inhibitors of COX-2 can decrease the growth of certain human tumors[25,26] and prevent tumorigenesis in animal models[27]. A pathological study showed increased COX-2 expression levels in gastric cancer[28,29]. Reduced COX-2 expression in gastric cancer cells led to markedly decreased proliferation and metastatic capability, demonstrating that COX-2 activity is necessary for gastric cancer cell proliferation and metastasis[30]. All of the above evidence indicates that COX-2 plays an important role in gastric cancer. In this study, we investigated the roles of miR-143 and COX-2 in gastric cancer and found that both miR-143-5p and miR-143-3p function as tumor suppressors in gastric cancer; however, miR-143-5p alone directly targeted COX-2 and exhibited a stronger tumor suppressive effect than miR-143-3p.
The human gastric cancer cell lines MKN-1, MKN-7, AGS, SGC-7901 and BGC-823 and the normal gastric epithelium cell line GES-1 were grown in RPMI 1640 medium supplemented with 10% FBS (Hyclone). The cell cultures were incubated in room air at 37 °C in a humidified atmosphere of 5% CO2.
Total RNA was extracted with TRIzol (Invitrogen). For mature miRNA expression analysis, cDNA was synthesized with the Taqman MiRNA Reverse Transcription kit (Applied Biosystems) and 100 ng of total RNA (100 ng/μL), along with 1 μL of miR-143-5p (Applied Biosystems) or miR-143-3p (Applied Biosystems) specific primers that were supplied with the miRNA Taqman MicroRNA Assay, according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (PCR) analyses were performed in triplicate on a 7900HT Real-Time PCR System (Applied Biosystems), and the data were normalized to RNU6B (Applied Biosystems) for each reaction. The thermal cycling profile used was as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Quantification was performed according to the standard ΔΔCT method.
Cells were seeded 24 h prior to transfection into 24-well or 6-well plates or 6 cm dishes. Hsa-miR-143-5p (Applied Biosystems), hsa-miR-143-3p (Applied Biosystems) or a miRNA mimic control (Applied Biosystems) was transfected with Lipofectamine 2000 (Invitrogen) at a final concentration of 50 nmol/L. The sequences of the mature miR-143-5p and miR-143-3p used in this study were GGUGCAGUGCUGCAUCUCUGGU and UGAGAUGAAGCACUGUAGCUC, respectively. The cells were harvested at 24 h (for RNA extraction), 48 h (for protein extraction) or 72 h (for apoptosis assays).
An Alamar blue assay was used to measure cell proliferation. This assay is based on the quantitative metabolic conversion of blue, non-fluorescent resazurin to pink, fluorescent resorufin by living cells. After 72 h of incubation, an Alamar blue (Invitrogen) stock solution was aseptically added to the wells to equal to 10% of the total incubation volume. The resazurin reduction in the cultures was determined after a 2-6 h incubation with Alamar blue by measuring the absorbances at 530-nm and 590-nm wavelengths on a Synergy HT Multi-Mode Microplate Reader (Bio-tek Instruments).
Following maintenance in culture, the cells were harvested and stained with phycoerythrin-conjugated Annexin V according to the manufacturer’s instructions (BD Biosciences). The cells were then analyzed on a FACSCalibur flow cytometer (BD Biosciences). The cells were considered viable if double negative, early apoptotic if positive for Annexin V alone and necrotic or late apoptotic if double positive.
RNA22 (http://cbcsrv.watson.ibm.com/rna22.html) was used to identify miRNA-target sites in the 3′-UTR of COX-2 mRNA and the corresponding RNA/RNA complexes and folding energies.
Cells were lysed with Radio Immunoprecipitation Assay buffer (Sigma-Aldrich), and the total protein concentration was determined with a Bio-Rad Protein Assay (Bio-Rad). Proteins (40 μg) were separated by 10% SDS/PAGE and electrotransferred onto nitrocellulose membranes. The membranes were then incubated overnight with a COX-2 (Cell Signaling) or poly (ADP-ribose) polymerase (PARP) primary antibody (Cell Signaling) at 4 °C, and subsequently incubated with an HRP-conjugated anti-rabbit secondary antibody (Bio-Rad) for 1 h at room temperature. Protein bands were detected with the Western Blotting Luminol Reagent (Santa Cruz Biotechnology).
Total RNA was extracted with TRIzol (Invitrogen). DNase I (Amplification Grade, Invitrogen) and the SuperScript First-Strand Synthesis System for reverse transcription-PCR (RT-PCR) (Invitrogen) were used for cDNA preparation. Primers and probes were ordered from IDT Inc. The following primers and probes were used: COX-2: Primer-F: 5’-CAAATCCTTGCTGTTCCCACCCAT-3’, Primer-R: 5’-GTGCACTGTGTTTGGAGTGGGTTT-3’, Probe: 5’-AAGTGCGATTGTACCCGGACAGGATT-3’, β-GUS: Primer-F: 5’-CTCATTTGGAATTTTGCCGATT-3’, Primer-R: 5’-CCGAGTGAAGATCCCCTTTTTA-3’, Probe: 5’-TGAACAGTCACCGACGAGAGTGCTGG-3’. The reactions were incubated in a 96-well plate at 95 °C for 12 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 45 s. All reactions were performed in triplicate.
The human COX-2 3′-UTR was amplified and cloned into the XbaI site of the pGL3-control vector (Promega, United States), downstream of the luciferase gene, to generate the plasmid pGL3-WT-COX2-3′-UTR. pGL3-MUT-5p-COX2-3′-UTR was generated from pGL3-WT- COX2-3′-UTR by deleting the “ACTGTAC” binding site for miR-143-5p. For the luciferase reporter assay, cells were cotransfected with the luciferase reporter vectors and miR-143-5p, miR-143-3p or a miRNA mimic control, using Lipofectamine 2000. A β-actin promoter Renilla luciferase reporter was used for normalization. After 48 h, luciferase activity was analyzed by the Dual-Glo Luciferase Assay System (Promega), according to the manufacturer’s protocols.
Experimental results were assessed for significance with one-tailed unpaired t tests. A P value less than 0.05 was considered significant.
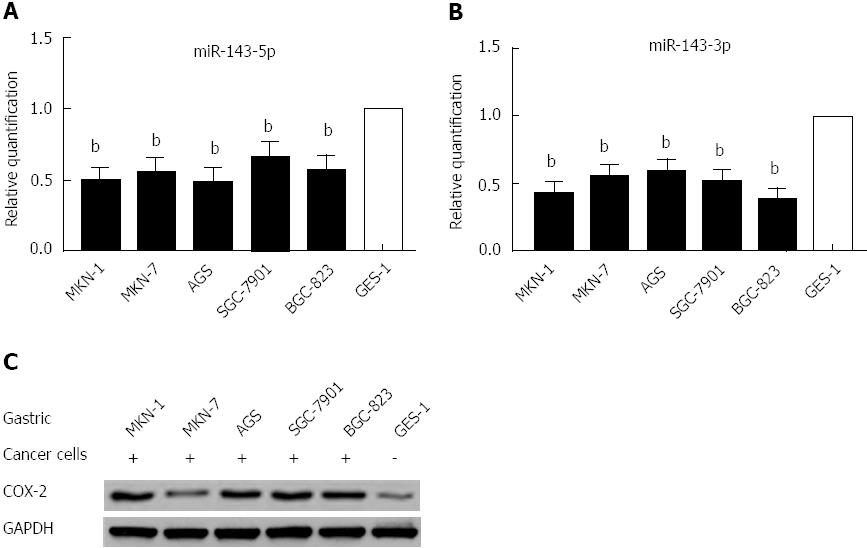
Using real-time PCR to quantify mature miR-143-5p and miR-143-3p in five human gastric cancer cell lines and a normal gastric epithelium cell line, we found that both the miR-143-5p and miR-143-3p expression levels were markedly reduced in gastric cancer cells (P < 0.01; Figure 1A and B). Western blot analysis indicated that COX-2 protein expression was increased in the five human gastric cancer cell lines (Figure 1C); this expression was inversely correlated with the miR-143 levels.
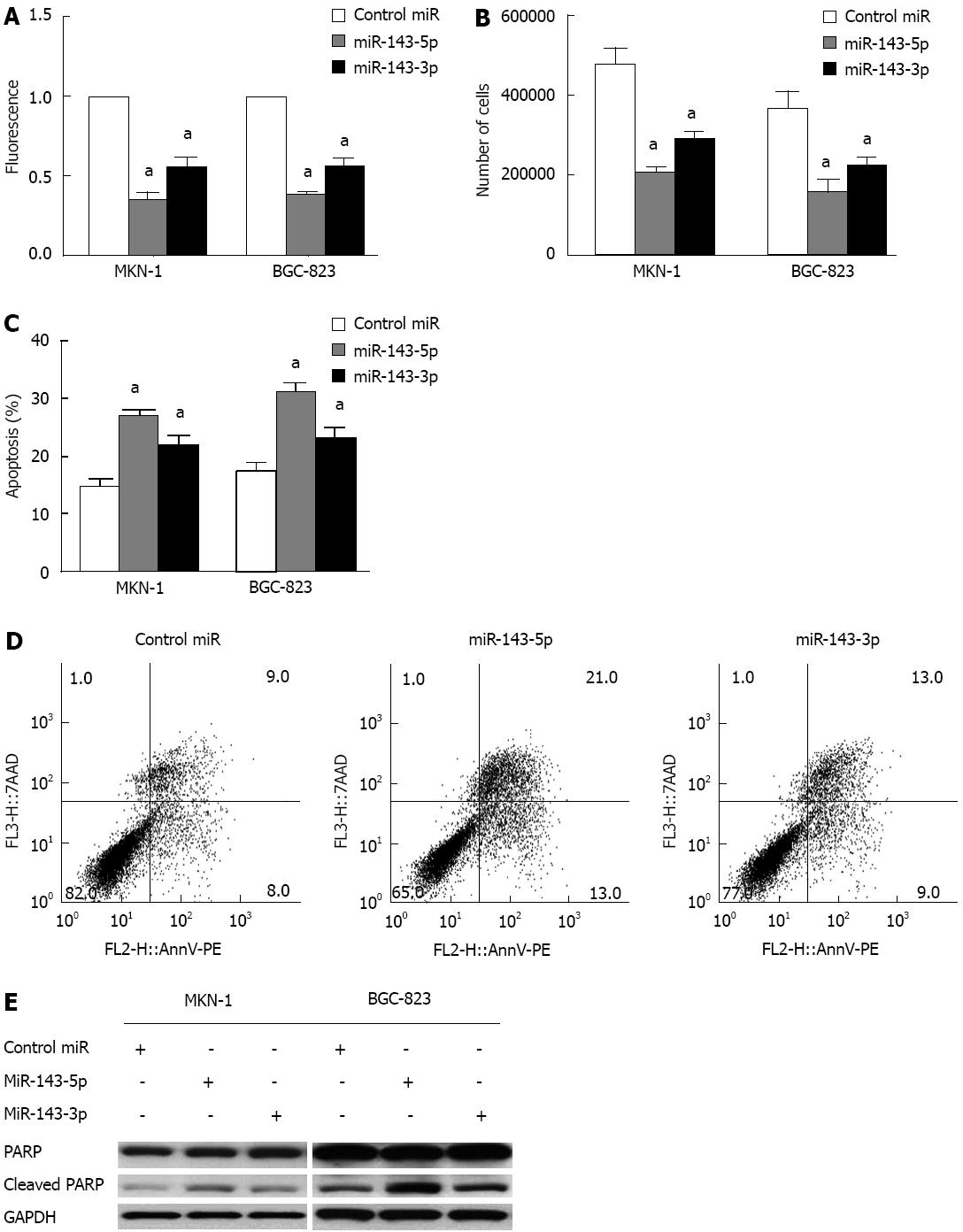
The reduced miR-143 expression in gastric cancer suggests that this miRNA could have anti-proliferative effects. To test this hypothesis, we evaluated the effects of transient transfection with miR-143-5p and miR-143-3p in MKN-1 and BGC-823 gastric cancer cell lines. The Alamar Blue assay, a redox assay similar to the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, showed significant decreases in gastric cancer cell viability following the transfection of either miR-143-5p or miR-143-3p. The decrease in viability after miR-143-5p transfection was greater than that after miR-143-3p transfection (P < 0.05; Figure 2A). Additionally, cell counts were significantly decreased after transfection with either miR-143-5p or miR-143-3p (P < 0.05; Figure 2B). Consistent with the results of the Alamar Blue assay, miR-143-5p showed a stronger inhibitory effect than miR-143-3p.
An apoptosis assay after transfection with miR-143-5p or miR-143-3p indicated a marked increase in cell apoptosis. Cells transfected with miR-143-5p had a significantly higher apoptosis rate than those transfected with miR-143-3p (P < 0.05; Figure 2C and D). Furthermore, both miR-143-5p and miR-143-3p transfection were accompanied by increased levels of cleaved PARP, a product of apoptosis (Figure 2E).
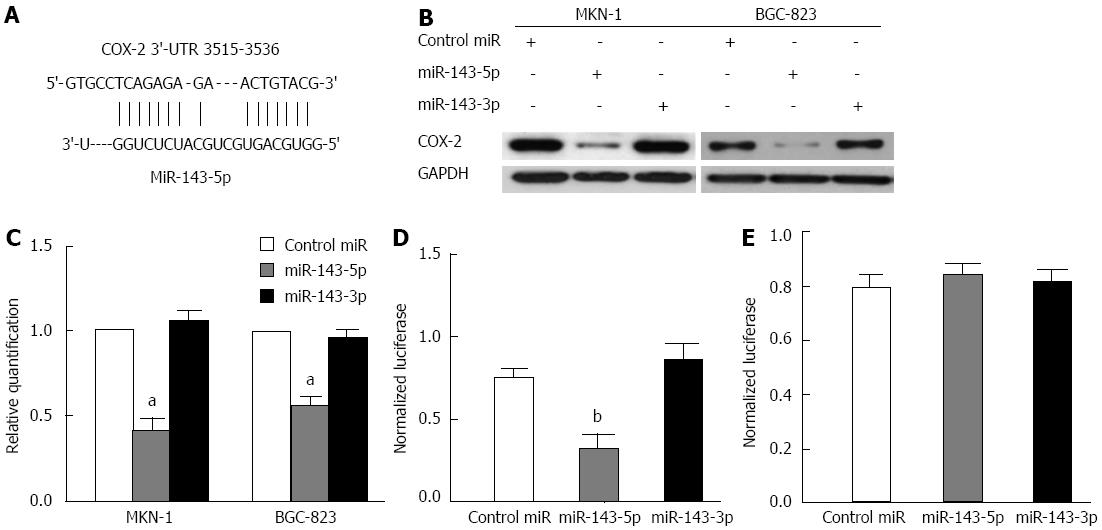
A bioinformatics analysis, conducted with RNA22, indicated that the 3′-UTR of the human COX-2 mRNA (NM_000963) harbors a putative miR-143-5p binding site (nucleotides 3515-3536) (Figure 3A) but has no miR-143-3p binding site. To assess the inhibitory effects of miR-143 on COX-2, MKN-1 and BGC-823 cells were transfected with miR-143-5p, miR-143-3p or a control. Western blot analysis revealed a profound decrease in the COX-2 protein level after transfection with miR-143-5p but not miR-143-3p (Figure 3B). Consistent with transcriptional inhibition by miRNA, we noted decreased COX-2 mRNA levels after miR-143-5p transfection (P < 0.05; Figure 3C).
To confirm the direct action of miR-143 on the COX-2 3′-UTR, transient transfection experiments were performed with the 3′-UTR of COX-2, containing mutated or non-mutated putative miR-143-5p matching sites, downstream of the luciferase open reading frame. Transfection with miR-143-5p inhibited the normalized activity of the COX-2 3′-UTR reporter by 57% (P < 0.01), whereas MiR-143-3p had no effect on this reporter activity (Figure 3D). In contrast, co-transfection of the mutant reporter plasmid with either miR-143-5p or miR-143-3p had no effect on luciferase activity in the transfected cells (Figure 3E). These results demonstrated that only miR-143-5p bound to the seed sequence present in the 3′-UTR of human COX-2 mRNA to inhibit COX-2 expression.
In the present study, the expression levels of both miR-143-5p and miR-143-3p were found to be downregulated in gastric cancer. This finding is consistent with reports from other research groups[22,23]. In a recent report, the authors used real-time RT-PCR and chip assays to analyze 70 paired samples of gastric cancers and benign tissues[23]. The authors found that miR-143 was among the most strongly downregulated miRNAs in gastric cancers, compared to benign tissues. The miR-143 expression level was associated with gastric cancer progression and was more significantly reduced in stage IV cancers, compared with stage I and II cancers. Consistent with our observations, a study by Takagi et al[22] also indicated that miR-143 was downregulated in gastric cancer cell lines and that transfection with miR-143-3p inhibited the gastric cancer cell viability by targeting ERK5 and AKT. However, the function of miR-143-5p has never been investigated in gastric cancer.
In the miRNA biogenesis pathway, long primary transcripts (pre-miRNAs) that have been transcribed from the genome are processed by the cellular RNase enzyme III Drosha into 60-110-nt structures called precursor miRNAs (pre-miRNA)[30]. Pre-miRNAs are cleaved by the RNase III enzyme Dicer-1 to produce short, imperfect, double-stranded miRNA duplexes that are subsequently unwound by helicases to create mature miRNAs. In some cases, two mature miRNAs can be excised from the same stem-loop pre-miRNA[31]. These “5p” and “3p” miRNAs, although generated from a single primary transcript, have different sequences and therefore target different mRNAs. In humans, two different mature miRNA sequences are excised from opposite arms of the stem-loop pre-miR-143 to generate two different miRNAs, hsa-miR-143-5p and has-miR-143-3p. Despite nearly a decade of studies on the roles of miRNA in cancers, the comparative roles of strand-specific mature miRNAs that originate from the same stem-loop precursor (5p and 3p) have yet to be fully studied. Previously, a number of studies demonstrated that ectopic miR-143 expression inhibited cancer cells in various types of human tumors[4,6-8,16,20,22]. However, the role of miR-143 in gastric cancer has not been fully investigated, and differences between miR-143-5p and miR-143-3p have never been reported.
Our functional analysis of miR-143 expression in gastric cancer cell lines indicated tumor suppressor functions for both miR-143-5p and miR-143-3p. Our data revealed that the restoration of both miR-143-5p and miR-143-3p expression suppressed cell proliferation and promoted apoptosis in gastric cancer cells. The tumor-suppressive effect of miR-143-5p was stronger than that of miR-143-3p.
Further analysis demonstrated that only miR-143-5p directly binds to COX-2 mRNA. The luciferase reporter assay revealed that COX-2 contains a binding site for miR-143-5p but no binding site for miR-143-3p. A recent report also demonstrated different functions of miR-28-5p and miR-28-3p; specifically, miR-28-5p altered the expression of CCND1 and HOXB3, whereas miR-28-3p bound to NM23-H1 in colorectal cancer[32]. Our study provides further evidence that strand-specific “5p” and “3p” miRNAs could have different targets. This could explain why miR-143-5p is a stronger tumor suppressor in gastric cancer and suggests the existence of other potential targets of miR-143-3p. For example, other studies have revealed multiple targets for miR-143, such as KRAS[5,6], Bcl-2[7,8], PAI-1[9], MMP-13[11], DNMT3A[12], ELK1[13] and MYO6[10]. Further studies are needed to determine other targets of miR-143-5p and miR-143-3p in gastric cancer.
In conclusion, both miR-143-5p and miR-143-3p are downregulated in gastric cancer and function as anti-oncomirs. MiR-143-5p is more strongly tumor suppressive than miR-143-3p. Our data also indicate that COX-2 is a direct target of miR-143-5p but not of miR-143-3p. Further studies are needed to define the detailed mechanisms and identify more miR-143 targets.
MicroRNA-143 (miR-143) is considered a pivotal regulator of gene expression and directly targets multiple mRNAs that code for proteins involved in cell proliferation, differentiation, survival and apoptosis. It is downregulated in various human tumors and suppresses tumor growth in cancers of the urogenital system, digestive system, respiratory system and nervous system, as well as some sarcomas and B-cell malignancies. MiR-143 expression has also been reported to be downregulated in human gastric cancer tissues and cell lines, but the mechanism by which miR-143 regulates cancer cells is not fully clear.
MiR-143 has been reported to be a tumor suppressor. However, the role of miR-143 in gastric cancer has not been fully investigated. The functional differences between miR-143-5p and miR-143-3p with regard to cancer have never been reported. In this study, the authors compared the tumor suppressive functions of miR-143-5p and miR-143-3p and explored the associated underlying mechanism.
This functional analysis of miR-143 expression in gastric cancer cell lines indicated that both miR-143-5p and miR-143-3p act as tumor suppressors. The restoration of either miR-143-5p or miR-143-3p suppressed cell proliferation and promoted apoptosis in gastric cancer cells. The tumor-suppressive effect of miR-143-5p was stronger than that of miR-143-3p. Further analysis demonstrated that only miR-143-5p directly bound to the cyclooxygenase-2 (COX-2) mRNA. The luciferase reporter assay revealed that COX-2 contained a binding site for miR-143-5p but not for miR-143-3p. Western blot showed a profound decrease in COX-2 protein expression after transfection with miR-143-5p but not with miR-143-3p. The data in this article indicate that COX-2 is a direct target of miR-143-5p but not of miR-143-3p.
Both miR-143-5p and miR-143-3p function as anti-oncomirs and are downregulated in gastric cancers. MiR-143-5p is more strongly tumor suppressive than miR-143-3p and could be a potential gastric cancer therapeutic target.
MiR-143 is considered a pivotal regulator of gene expression because it directly targets multiple mRNAs that code for proteins involved in cell proliferation, differentiation, survival and apoptosis. It is downregulated in various human tumors and suppresses tumor growth in cancers from the urogenital, digestive, respiratory and nervous system, as well as B-cell malignancies.
The authors examined the effects and mechanisms of miR-143 subtypes on gastric cancer cell lines. They examined the effects of these oncomirs on growth, apoptosis and COX-2 activity. For the most part, the paper is straightforward and well written. The experiments are well described and the results are clearly presented.
P- Reviewers: Hahm KB, Kanda T, Raffaniello RD S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Ma S
| 1. | Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2967] [Cited by in RCA: 2888] [Article Influence: 169.9] [Reference Citation Analysis (0)] |
| 2. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2135] [Cited by in RCA: 2103] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 3. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 4. | Song T, Zhang X, Wang C, Wu Y, Dong J, Gao J, Cai W, Hong B. Expression of miR-143 reduces growth and migration of human bladder carcinoma cells by targeting cyclooxygenase-2. Asian Pac J Cancer Prev. 2011;12:929-933. [PubMed] |
| 5. | Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 450] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 6. | Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z, Li P, Zhang W, Wu H, Feng N. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 7. | Liu L, Yu X, Guo X, Tian Z, Su M, Long Y, Huang C, Zhou F, Liu M, Wu X. miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Report. 2012;5:753-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 8. | Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24:1363-1369. [PubMed] |
| 9. | Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M, Ørntoft TF. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012;106:366-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 10. | Szczyrba J, Löprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 11. | Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 217] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 12. | Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, Röcken C, Ebert MP, Kwok TT, Sung JJ. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101:699-706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 223] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 13. | Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705-710. [PubMed] |
| 14. | Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ, Kerin MJ. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF, Huang Q, Fang GQ, Li EM. MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J Gastroenterol. 2011;17:79-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 77] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, Chang YS, Chen SJ. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 236] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 18. | Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, Shu Y. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 203] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 19. | Amaral FC, Torres N, Saggioro F, Neder L, Machado HR, Silva WA, Moreira AC, Castro M. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metab. 2009;94:320-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 127] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 20. | Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL, Khanin R, O’Connor R, Mihailovic A, Taylor BS. Small RNA sequencing and functional characterization reveals MicroRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011;71:5659-5669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 21. | Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 222] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 22. | Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 242] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 23. | Li X, Luo F, Li Q, Xu M, Feng D, Zhang G, Wu W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep. 2011;26:1431-1439. [PubMed] |
| 24. | Hoellen F, Kelling K, Dittmer C, Diedrich K, Friedrich M, Thill M. Impact of cyclooxygenase-2 in breast cancer. Anticancer Res. 2011;31:4359-4367. [PubMed] |
| 25. | Adhim Z, Matsuoka T, Bito T, Shigemura K, Lee KM, Kawabata M, Fujisawa M, Nibu K, Shirakawa T. In vitro and in vivo inhibitory effect of three Cox-2 inhibitors and epithelial-to-mesenchymal transition in human bladder cancer cell lines. Br J Cancer. 2011;105:393-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Yin J, Liu B, Li B, Liu Z, Xie X, Lv Z, Gao S, Guang J. The cyclooxygenase-2 inhibitor celecoxib attenuates hepatocellular carcinoma growth and c-Met expression in an orthotopic mouse model. Oncol Res. 2011;19:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Zhong B, Cai X, Chennamaneni S, Yi X, Liu L, Pink JJ, Dowlati A, Xu Y, Zhou A, Su B. From COX-2 inhibitor nimesulide to potent anti-cancer agent: synthesis, in vitro, in vivo and pharmacokinetic evaluation. Eur J Med Chem. 2012;47:432-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Ma D, Liu M, Wang AP, Yang H. Cycloxygenase-2 is essential for the survival and proliferation of gastric cancer cells. Cell Biochem Biophys. 2011;61:637-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Wu WK, Cho CH, Lee CW, Fan D, Wu K, Yu J, Sung JJ. Dysregulation of cellular signaling in gastric cancer. Cancer Lett. 2010;295:144-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 30. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3629] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 31. | Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-D144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3282] [Cited by in RCA: 3542] [Article Influence: 186.4] [Reference Citation Analysis (0)] |
| 32. | Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886-896.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |