Published online Jan 28, 2013. doi: 10.3748/wjg.v19.i4.440
Revised: October 26, 2012
Accepted: December 15, 2012
Published online: January 28, 2013
A recent work of Iliopoulos et al published in Cell highlighted a circuit orchestrated by microRNAs (miRNAs) that results in liver tumorigenesis and inflammation. This feedback loop, governed by miR-24 and miR-629, promotes a hepatocyte nuclear factor-4α transient inhibition resulting in miR-124 induction and signal transducer and activator of transcription 3 activation. These promising data support the use of miRNA mimics or inhibitors as potent therapeutic approaches in liver cancer.
- Citation: Gougelet A, Colnot S. MicroRNA-feedback loop as a key modulator of liver tumorigenesis and inflammation. World J Gastroenterol 2013; 19(4): 440-444
- URL: https://www.wjgnet.com/1007-9327/full/v19/i4/440.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i4.440
Hepatocyte nuclear factor-4α (HNF-4α) belongs to the nuclear receptor superfamily of ligand dependent transcription factors NR2A1[1]. It is expressed in the kidney, intestine and pancreas. HNF-4α is most highly expressed in the liver[2-5], where it binds to the promoter of 12% of genes expressed in the adult liver[6]. It plays a key role in liver development during gastrulation[7] and orchestrates the parenchyma formation in the adult liver[8]. It also guides hepatoblasts to hepatocyte differentiation[9]. The conditional disruption of HNF-4α in mouse liver showed that HNF-4α regulates a number of genes involved in lipid, glucose, amino acid and xenobiotic metabolisms[4]. Related to its central role in liver physiology, HNF-4α has been reported to be involved in liver oncogenesis; however, its role is still controversial. A study by Xu et al[10] suggested that HNF-4α expression was increased in human hepatocarcinoma (HCC), which contrasted with other studies that reported that the HNF-4α level was decreased in HCC[11-13]. HNF-4α has also been associated with cancer and inflammation in the colon: its intestinal disruption in mouse revealed that HNF-4α protects against colitis and inflammatory disease[14,15] and promotes tumorigenesis[15]. Liver cancer is a clear example of an inflammation-related cancer[16-19]; therefore, it does not seem surprising that HNF-4α could also orchestrate an inflammatory program in the liver, as was shown last year by Wang et al[20] in experiments performed on hepatocarcinoma cells exposed to interleukin (IL)-6.
In a recent paper published in Cell, D. Iliopoulos’ Laboratory aimed to decipher the epigenetic circuit centered on HNF-4α, which promotes liver oncogenesis[21]. They particularly focused on microRNAs (miRNAs), which are small RNAs produced from non-coding DNA regions that control gene expression by binding to the 3’ untranslated region (3’ UTR) of target messenger RNAs (mRNAs). miRNA binding results in the degradation of the mRNA and/or the repression of its translation[22]. Since their discovery in 1994 in Caenorhabditis elegans[23], miRNAs have become an expanding area of research, and evidence supports the view that miRNAs are key regulators of many physiological processes, e.g., growth, proliferation and differentiation[24-27]. They are also implicated in the initiation and progression of various cancers[28-30]. Concerning HCC, microtranscriptomic analyses revealed a miRNA signature of liver cancer, i.e., miR-122 loss[31] or miR-221 upregulation[32], and a number of studies support the view that miRNAs direct HCC progression through the induction of cell proliferation or metastasis and the suppression of apoptosis[33]. Taken together, these data highlight miRNAs as promising diagnostic and therapeutic targets for HCC.
The work of Maria Hatziapostolou is a robust description of the molecular factors underlying HNF-4α loss-mediated hepatocarcinoma based on in vitro and in vivo experiments and confirmed on HCC human samples. Prior to the precise deciphering of hepatocarcinoma molecular origins, the authors confirmed that the silencing of HNF-4α was sufficient to increase cell colony formation and invasion of nontransformed immortalized human hepatocytes (IMH) and human liver cancer cell lines. On their own, the injection of IMH cells with a preliminary transient inhibition of HNF-4α in immunosuppressed mice resulted in tumor apparition. Interestingly, HNF-4α was maintained at a low level 55 d after cell xenografts, suggesting that transient silencing of HNF-4α initiated a feedback loop that stably maintained cell transformation and suppressed its own expression.
What are the factors involved in HNF-4α silencing? An miRNA screening was performed on a luciferase reporter gene containing the 3’ UTR region of HNF-4α, which revealed that miR-24 and, to a lesser extent, miR-629, were HNF-4α repressors. The transient expression of both miRNAs, the equivalent of HNF-4α silencing, transformed IMH cells, increased their invasiveness in vitro and promoted the appearance of tumors in mice. Strikingly, both miRNAs were induced following HNF-4α silencing, supporting the involvement of these miRNAs in the HNF-4α- dependent feedback loop.
Interestingly, miR-24 and miR-629 possess a binding motif for signal transducer and activator of transcription 3 (STAT3) in their promoters, a well-known effector of IL-6-dependent cancer inflammation[34] (for a review), particularly in liver cancer[35] (for a review). In brief, the binding of IL-6 to its receptor, IL-6R, activates the Janus kinase family, which phosphorylates STAT3 on tyrosine residues. STAT3 could then form homodimers before translocation into the nucleus to promote the expression of an inflammatory program[36] (for a review). Here, the authors showed that in response to IL-6, STAT3 binding to miR-24 and miR-629 promoters increased, resulting in miRNA expression. In turn, miR-24 expression and HNF-4α silencing enhanced the phosphorylation status of STAT3, and thus its activation. STAT3 and IL-6 are two other members of the HNF-4α feedback circuit promoting liver oncogenesis; an antibody against IL-6 blocked all the HNF-4α silencing effects that they observed.
The next step consisted in screening of miRNAs possessing an HNF-4α binding site, which identified miR-124 as a preferential HNF-4α target. miR-124 was, in fact, an integral part of the feedback loop, because its inhibition by an antisense as-miR-124, or following HNF-4α silencing, enhanced STAT3 phosphorylation coupled to an induction of IL-6 secretion and expression of IL-6R, a direct target of miR-124.
The relevance of the loop miR-24/miR-629/HNF-4α/miR-124/STAT3 was confirmed in vivo with a mouse model of HCC induced by diethylnitrosamine. During tumor progression, the HNF-4α level was initially dropped off after four weeks, followed by miR-124 repression and inversely by miR-24 and IL-6R induction, which supports the essential role of the HNF-4α loop in tumor progression. One of the important finding of this study was that an injection of miR-124-mimic encapsulated into liposomes in this model was sufficient to limit tumor growth, even if this mimic was administered four weeks before analysis. miR-124 mimics were also able to prevent tumor development. Taken together, these results argue in favor of miR-124 delivery as a potent strategy for treating or preventing HCC. Although, to date, the efficacy of miRNA mimics have not been demonstrated in vivo, this approach could be attractive. The prerequisite for this approach is that the complementary passenger RNA in the double stranded miRNA mimic does not create a new miRNA that could induce off-target effects[37,38].
The second exciting idea from this study relies upon the need for an inflammatory context to optimally disrupt the HNF-4α epigenetic circuit and promote tumorigenesis. If the tumors developed in a mouse model lacked STAT-3 in the liver, the tumors were smaller when the HNF-4α loop was less inhibited. This supports the hypothesis that HNF-4α loss engages an anti-tumorigenic program and a STAT3-dependent anti-inflammatory program, both orchestrated by the trio miR-24/miR-629/miR-124, to induce hepatocyte transformation. Such an epigenetic loop has already been described by the same authors for breast cancers, in which cell transformation was dictated by an epigenetic switch achieved by IL-6, nuclear factor-κB and the miRNA let-7[39].
Finally, the authors demonstrated the relevance of their model for human liver oncogenesis: more than 50% of HCC samples from patients showed a loss of HNF-4α and of miR-124, accompanied by an increase in miR-24 and IL-6R expression; all these parameters were associated with a higher level of phospho-STAT3 status in relation with IL-6 and IL-6R expression. As a confirmation of the HNF-4α circuit’s key role in HCC progression, the authors showed that the activity of the HNF-4α circuit becomes more deregulated as the HCC grade increased.
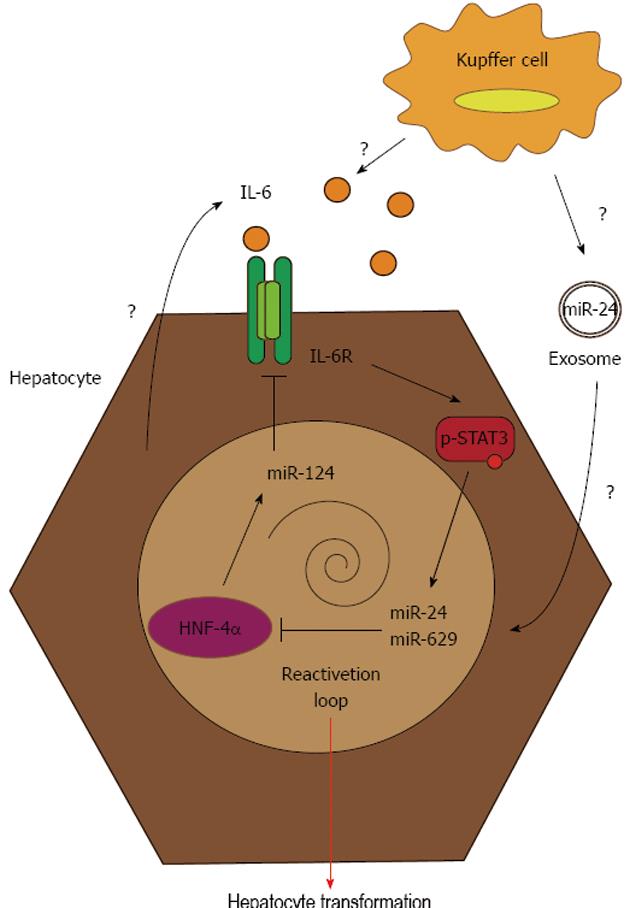
In summary, this work strongly argues for a tumor suppressor role of HNF-4α in liver, as was already shown[12], and supports the key role of this transcription factor in limiting the tumor inflammatory environment by inhibiting the IL-6/STAT3 pathway. The authors described the interconnection between the trio miR-24/miR-629/miR-124 and HNF-4α as a key regulator of HCC progression (Figure 1). In 2010, Takagi et al[40] described a crosstalk between HNF-4α and miR-24 in a liver metabolic loop for the control of bile acid synthesis. In brief, bile acids induce reactive oxygen species generation, resulting in the activation of the mitogen-activated protein (MAP) kinase pathway. In turn, miR-24, induced by the MAP kinases, downregulates HNF-4α, and, thus, the expression of bile acid-synthesizing enzymes. Hepatitis B virus (HBV) and alcohol abuse, two etiological agents associated with HCC development, contribute to oxidative stress in the liver. In consequence, miR-24 inhibition and, thus, reactivation of the HNF-4α feedback circuit in HCC could also be performed using antioxidants, a class of therapeutic agents currently used for HBV patients[41].
The take-home message of this study is that a transient signal at any level of the feedback circuit (HNF-4α or miR-124 silencing, miR-24 overexpression or IL-6 secretion) stably induces hepatocyte transformation, which could be auto-amplified by this feedback loop, and lead to chronic inflammation, predisposing a patient to liver cancer development and immune escape[19]. The activation of the IL-6/STAT3 axis is, at least in part, caused by a hepatocyte source, as shown by the in vitro experiments, but this could also be boosted by IL-6 secretion by non-parenchymal liver cells like Kupffer cells. Another possible scenario for the disruption of HNF-4α circuit by tumor environment could be the secretion of exosomes containing miRNA, in particular miR-24, by immune cells. Exosomes are small extracellular vesicles derived from the multivesicular body-sorting pathway, which are produced by various cells like epithelial cells, immune cells and tumor cells. Recently, it has been shown that some miRNAs, particularly those deregulated in cancer, are encapsulated into exosomes and play a role as cell-cell mediators[41]. In particular, miR-24 has been detected in exosomal structures, and a similar mechanism might exist in liver, permitting miR-24 to be delivered to hepatocytes by neighboring cells.
The most promising result of this study was that intravenous delivery of miR-124 mimic encapsulated into liposomes was able to prevent and reduce the growth of liver cancer. To date, few works have demonstrated the potency of an miRNA-based anticancer approach. However, the great advantage of HCC is that mimics or inhibitors against miRNAs are preferentially distributed to the liver[42]. For that reason, the first miRNA-based therapy authorized in clinical trials is the anti-miR-122 compound Miravirsen, indicated for the treatment of hepatitis C virus (HCV), which uses miR-122 in hepatocytes for its replication. This clinical trial follows promising results obtained in primates infected with HCV and treated with an anti-miR-122[43], which was well-tolerated[44]. Concerning the HNF-4α circuit, the authors showed the efficiency of a mimic of miR-124 to perturb HCC development, but we could also conceive the injection of inhibitors against miR-24 and miR-629 to avoid HNF-4α loss. Even if miRNA inhibitor delivery appeared as a promising therapeutic approach, the use of these modulators poses a number of challenges. The question of treatment specificity could be raised. In fact, an miRNA could target multiple mRNAs[45,46], and, thus, many cellular pathways, pro- or anti-tumorigenic. It could be difficult to restrict the effect of one miRNA to only one target of interest. However, this notion could be generalized to many other treatments currently used, such as proteasome[47] or histone deacetylase inhibitors[48], which have demonstrated efficacy. Another major disadvantage could be the inhibition of a non-identified target. Their delivery could also be an important challenge, although many improvements have appeared recently, with chemical modifications limiting their degradation by nucleases[49-51]. The other problem is that these inhibitors preferentially target the liver and other tissues are more difficult to enter[42]. Finally, these inhibitors could also be toxic[52,53]. As suggested by the data of Hatziapostolou, mimics or inhibitors could be delivered in a free formulation, or protected into liposomes or even into exosomes, as was shown for siRNAs[54]. In conclusion, miR-124-based treatment could be a therapeutic strategy of choice for HCC, as an alternative to the anti-STAT3 approach[55]. This therapeutic option has the great advantage of targeting the inflammatory, as well as the metabolic initiators, of HCC, and its efficiency demonstrated in this work argue for an epigenetic switch as the major event underlying cancer initiation.
P- Reviewer Bonino F S- Editor Gou SX L- Editor Stewart GJ E- Editor Xiong L
| 1. | Sladek R, Giguère V. Orphan nuclear receptors: an emerging family of metabolic regulators. Adv Pharmacol. 2000;47:23-87. [PubMed] |
| 2. | Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207-1220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 3. | Gupta RK, Gao N, Gorski RK, White P, Hardy OT, Rafiq K, Brestelli JE, Chen G, Stoeckert CJ, Kaestner KH. Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev. 2007;21:756-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 135] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 4. | Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 888] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 5. | Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritié L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1027] [Cited by in RCA: 1033] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 7. | Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466-2477. [PubMed] |
| 8. | Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 479] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 9. | Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 337] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 10. | Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y, Ji Y, Wu X, Han Z, Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176-3181. [PubMed] |
| 11. | Lazarevich NL, Fleishman DI. Tissue-specific transcription factors in progression of epithelial tumors. Biochemistry (Mosc). 2008;73:573-591. [PubMed] |
| 12. | Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640-7651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 13. | Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908-920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 15. | Darsigny M, Babeu JP, Seidman EG, Gendron FP, Levy E, Carrier J, Perreault N, Boudreau F. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 2010;70:9423-9433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Anson M, Crain-Denoyelle AM, Baud V, Chereau F, Gougelet A, Terris B, Yamagoe S, Colnot S, Viguier M, Perret C. Oncogenic β-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. J Clin Invest. 2012;122:586-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 17. | Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 492] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 18. | Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 910] [Cited by in RCA: 954] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 19. | Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Ann NY Acad Sci. 2009;1155:206-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 294] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 20. | Wang Z, Bishop EP, Burke PA. Expression profile analysis of the inflammatory response regulated by hepatocyte nuclear factor 4α. BMC Genomics. 2011;12:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233-1247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 387] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 22. | Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 845] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 23. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 24. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4964] [Cited by in RCA: 5313] [Article Influence: 253.0] [Reference Citation Analysis (0)] |
| 25. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2105] [Cited by in RCA: 2355] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 26. | Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 798] [Cited by in RCA: 913] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 27. | Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1065] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 28. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6028] [Article Influence: 317.3] [Reference Citation Analysis (0)] |
| 29. | Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643-2646. [PubMed] |
| 30. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 31. | Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526-3536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 585] [Cited by in RCA: 590] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 32. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 613] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 33. | Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 34. | Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2935] [Cited by in RCA: 3389] [Article Influence: 211.8] [Reference Citation Analysis (0)] |
| 35. | He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 879] [Cited by in RCA: 948] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 36. | Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2296] [Cited by in RCA: 2404] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 37. | van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919-928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 319] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 38. | Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res. 2011;28:2996-3015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 156] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 39. | Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1167] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 40. | Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415-4422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 130] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 41. | Ha HL, Shin HJ, Feitelson MA, Yu DY. Oxidative stress and antioxidants in hepatic pathogenesis. World J Gastroenterol. 2010;16:6035-6043. [PubMed] |
| 42. | Roberts J, Palma E, Sazani P, Ørum H, Cho M, Kole R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol Ther. 2006;14:471-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1291] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 44. | Hildebrandt-Eriksen ES, Aarup V, Persson R, Hansen HF, Munk ME, Ørum H. A locked nucleic acid oligonucleotide targeting microRNA 122 is well-tolerated in cynomolgus monkeys. Nucleic Acid Ther. 2012;22:152-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 45. | Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2967] [Cited by in RCA: 2888] [Article Influence: 169.9] [Reference Citation Analysis (0)] |
| 46. | Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2577] [Cited by in RCA: 2717] [Article Influence: 159.8] [Reference Citation Analysis (0)] |
| 47. | Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood. 2009;113:4667-4676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 48. | Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 386] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 49. | Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1047] [Cited by in RCA: 960] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 50. | Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1659] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 51. | Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 390] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 52. | Agrawal S, Zhao Q. Mixed backbone oligonucleotides: improvement in oligonucleotide-induced toxicity in vivo. Antisense Nucleic Acid Drug Dev. 1998;8:135-139. [PubMed] |
| 53. | Zhou W, Agrawal S. Mixed-backbone oligonucleotides as second-generation antisense agents with reduced phosphorothioate-related side effects. Bioorg Med Chem Lett. 1998;8:3269-3274. [PubMed] |
| 54. | Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3883] [Cited by in RCA: 3569] [Article Influence: 254.9] [Reference Citation Analysis (0)] |
| 55. | Li WC, Ye SL, Sun RX, Liu YK, Tang ZY, Kim Y, Karras JG, Zhang H. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin Cancer Res. 2006;12:7140-7148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 117] [Article Influence: 6.5] [Reference Citation Analysis (0)] |