Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5813
Revised: April 30, 2013
Accepted: July 4, 2013
Published online: September 21, 2013
AIM: To study the subtype prevalence and the phylogenetic relatedness of hepatitis C virus (HCV) sequences obtained from the Argentine general population, a large cohort of individuals was analyzed.
METHODS: Healthy Argentinian volunteers (n = 6251) from 12 provinces representing all geographical regions of the country were studied. All parents or legal guardians of individuals younger than 18 years provided informed written consent for participation. The corresponding written permission from all municipal authorities was obtained from each city or town where subjects were to be included. HCV RNA reverse transcription-polymerase chain reaction products were sequenced and phylogenetically analyzed. The 5’ untranslated region (5’UTR) was used for RNA detection and initial genotype classification. The NS5B polymerase region, encompassing nt 8262-8610, was used for subtyping.
RESULTS: An unexpectedly low prevalence of HCV infection in the general population (0.32%) was observed. Our data contrasted with previous studies that reported rates ranging from 1.5% to 2.5%, mainly performed in selected populations of blood donors or vulnerable groups. The latter values are in keeping with the prevalence reported by the 2007 Argentinian HCV Consensus (approximately 2%). HCV subtypes were distributed as follows: 1a (25%), 1b (25%), 2c (25%), 3a (5%), and 2j (5%). Two isolates ascribed either to genotype 1 (5%) or to genotype 3 (5%) by 5’UTR phylogenetic analysis could not be subtyped. Subtype 1a sequences comprised a highly homogeneous population and clustered with United States sequences. Genotype 1b sequences represented a heterogeneous population, suggesting that this genotype might have been introduced from different sources. Most subtype 2c sequences clustered close to the 2c reported from Italy and Southern France.
CONCLUSION: HCV has a low prevalence of 0.32% in the studied general population of Argentina. The pattern of HCV introduction and transmission in Argentina appears to be a consequence of multiple events and different for each subtype.
Core tip: This study reports an unexpectedly low prevalence of hepatitis C virus (HCV) (0.32%) in the general population of Argentina, with a subgenotype distribution of 1a (25%), 1b (25%), 2c (25%), 3a (5%), and 2j (5%) while two isolates ascribed either to genotype 1 (5%) or to genotype 3 (5%) could not be subtyped. Phylogenetic analysis of the NS5B region has enabled us to define the pattern of HCV introduction and transmission in Argentina as a consequence of multiple events that differed for each (sub)genotype studied. Furthermore, this report discusses the putative sources of HCV subgenotype introduction in Argentina.
- Citation: del Pino N, Oubiña JR, Rodríguez-Frías F, Esteban JI, Buti M, Otero T, Gregori J, García-Cehic D, Camos S, Cubero M, Casillas R, Guàrdia J, Esteban R, Quer J. Molecular epidemiology and putative origin of hepatitis C virus in random volunteers from Argentina. World J Gastroenterol 2013; 19(35): 5813-5827
- URL: https://www.wjgnet.com/1007-9327/full/v19/i35/5813.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i35.5813
The analysis of extensive sets of sequences from hepatitis C virus (HCV) isolates throughout the world has revealed the existence of six major genetic groups or genotypes (named 1 to 6), and a large number of subtypes within these groups (designated a, b, c, etc.)[1]. Overall sequence divergence ranges from 31% to 33% among genotypes and from 20% to 25% among subtypes[1,2]; in a single patient, cloned E1/E2 sequences may differ by up to 10%. The reason for this great variation is a high mutation rate and high level of viral replication through an error prone RNA polymerase without proofreading capacity. Consequently, in the infected individual, the virus circulates as a complex viral quasispecies[3] whose composition is subject to continuous changes due to competitive selection[4] and interactions among variants of with different levels of fitness[5]. The calculated average rate of fixation of mutations has consistently been found to range between 1.1 and 1.5 × 103 mutations per site and per year[6,7]. The rate of fixation of mutations is not evenly distributed throughout the genome, which has highly variable regions within the envelope coding genes and well conserved regions, such as the 5’ untranslated region (5’UTR). The practical consequence of the high conservation at 5’UTR for HCV genotyping is that this region contains insufficient variation to solve HCV classification at the level of viral subtypes[8]. Furthermore, genotype 6 variants other than 6a and 6b show 5’UTR sequences identical or similar to those of type 1 and, therefore, cannot be differentiated[9-11]. It has been reported that sequence analysis of the highly conserved region in NS5B known as the “Okamoto region” (nt 8282 to 8610 in the H77 reference genome)[8] provides the best concordance with the full length genome phylogeny for precise genotype identification. The same primers can amplify genotypes 1 to 5, and they are less efficient for genotype 6 isolates, but they facilitate analysis and classification of the amplified sequences[11].
The prevalence of HCV infection in Argentina has been reported at 1.5% when all age groups are considered, and up to 2.0%-2.5% in adults[12], a rate in keeping with the value reported by the Argentinian Consensus on Hepatitis C (approximately 2%) in 2007. A higher prevalence (4.9%-5.7%) has been described in small rural communities[13,14]. HCV genotype distribution in Argentina in groups at risk of HCV infection (e.g., transfusion patients, hemodialysis patients, intravenous drug users, healthcare workers) has been reported at 53.5% for genotype 1, 23% for genotype 2 (mainly by subtype 2c [HCV-2c]), 8.6% for genotype 3, and 14.8% for mixed infections[15]. Similar results have been reported in studies on sequences deposited in GenBank (GB) and analyses by the Los Alamos database http://hcv.lanl.gov, with few genotype distribution changes (79.5% for genotype 1, 13.9% for genotype 2, 3.9% for genotype 3, 2.4% for genotype 4), but with some differences depending on the HCV subgroup at risk studied[16-18]. Phylogenetic characterization of genotype 4 isolates from Argentina has traced an independent origin of the three sequences studied[17]. Interestingly, HCV-2c was the most prevalent subtype (58%) in the city of Córdoba (Central geographical region of the country), followed by HCV-1b (33%), and to a lesser extent by HCV-1a (11%), HCV-3a (3%) and HCV-4a (3%)[19,20].
Here, we report an unexpectedly low prevalence of HCV infection (0.32%) as measured by anti-HCV antibodies detected by using both a second generation enzyme immune assay (EIA) and a confirmatory immunoblotting, and HCV RNA detected by reverse transcription - nested polymerase chain reaction (RT-nested PCR) targetting the 5’UTR HCV RNA in a cohort of random Argentinian volunteers. The genotypes detected and the putative origin of the HCV sequences are discussed based based on both their phylogenetic clustering and on such clustering relative to other Argentinian and worldwide derived sequences deposited in GB, in an attempt to trace how HCV could have been introduced in the local community here represented by the cohort studied.
Throughout the 2000-2007 period, a total of 6251 serum samples were collected from healthy volunteers from 12 Argentinian provinces, as well as from the Ciudad Autónoma de Buenos Aires (C.A.B.A. - the capital city of the country) as follows: Buenos Aires province and C.A.B.A., n = 1461; Catamarca, n = 648; Córdoba, n = 1061; Chaco, n = 353; Chubut, n = 172; Entre Ríos, n = 474; Jujuy, n = 176; Río Negro, n = 329; Salta, n = 561; San Luis, n = 195; Santiago del Estero, n = 375; and Tucumán, n = 446 (Table 1).
| Province/city of residence | Total number of individuals | Male/female number | Age1 (yr), mean ± SE |
| Buenos Aires/C.A.B.A. | 1461 | 685/776a | 33.4 ± 0.3 (11.2, 30)d |
| Catamarca | 648 | 267/381 | 39.3 ± 0.5 (13.0, 38)d |
| Córdoba | 1061 | 393/668b | 37.1 ± 0.4 (12.5, 35) |
| Chaco | 353 | 175/178b | 40.2 ± 0.8 (14.1, 40)d |
| Chubut | 172 | 83/89 | 37.4 ± 0.9 (12.1, 37) |
| Entre Ríos | 474 | 225/249 | 38.5 ± 0.5 (11.6, 38) |
| Jujuy | 176 | 105/71b | 35.2 ± 0.8 (10.3, 34)d |
| Río Negro | 329 | 149/180 | 40.1 ± 0.7 (12.8, 39)d |
| Salta | 561 | 230/331 | 41.3 ± 0.5 (12.8, 41)d |
| San Luis | 195 | 52/143b | 41.6 ± 1.2 (16.3, 40)d |
| Santiago del Estero | 375 | 164/211b | 38.0 ± 0.7 (13.2, 35) |
| Tucumán | 446 | 209/237 | 37.7 ± 0.6 (12.0, 35) |
| Total | 6251 | 2738/3513 | 37.5 ± 0.2 (12.7, 35) |
Subjects included in this study [n = 6251; 2738 men; mean ± SE, 37.5 ± 0.2 years; mean ± SD = 37.5 ± 12.7; median age = 35 years (range 10-70 years)] were recruited as volunteers from the general population, local schools, and police stations, after being informed about the aim of the survey. All parents or legal guardians of individuals younger than 18 years provided informed written consent for participation. The corresponding written permission from all municipal authorities was obtained from each city or town where subjects were to be included.
The presence of anti-HCV antibodies was determined by using a second generation EIA test according to the manufacturer’s recommendations (Abbott Diagnostics, North Chicago, IL, United States). Samples were further analyzed with a second generation recombinant immunoblot assay (RIBA 2.0: Chiron Corporation, Emeryville, CA, United States).
Samples with serologically detectable anti-HCV antibodies were subjected to either RT-nested or RT-hemi-nested PCR amplification (see below). The 5’UTR region was used for RNA detection and initial genotype classification. The NS5B polymerase region, encompassing nt 8262-8610, was used for subtyping.
RNA was extracted from 140 μL of serum by using the QIAamp Viral RNA Mini Kit (Qiagen Hilden, Germany). The measures to prevent contamination suggested by Kwok and Higuchi were strictly applied[21].
The 5’UTR RT-nested PCR was performed as follows. RT was carried out for 45 min at 42 °C (GeneAmp 2700 PCR system, Applied Biosystems, Foster City, CA, United States), using 50 U M-MLV reverse transcriptase, RNase H Minus, Point Mutant (200 U/μL Promega, Madison, WI, United States), 20 U RNase inhibitor (40 U/μL Promega, Madison, WI, United States), 10 mmol/L of each dNTP (Roche, Basel, Switzerland), 20 pmols of antisense PCR primer NR5 5’TGCTCATGGTGCACGGTCTACGAG3’ and 1 × buffer from the high fidelity Pfu turbo DNA polymerase (Stratagene, San Diego, CA, United States) in a final volume of 20 μL. Then, 80 μL of PCR mix containing 1 ×Pfu turbo buffer, 20 pmol of sense primer NF5 5’GTGAGGAACTACTGTCTTCACGCAG3’ and 2.5 U Pfu turbo DNA polymerase were added to each tube. After an initial denaturation step of 2 min at 95 °C, 5 initial cycles of 30 s at 94 °C, 30 s at 55 °C and 2 min at 72 °C were carried out, followed by 35 cycles of 30 s at 94 °C, 30 s at 60 °C and 2 min at 72 °C, finishing with a single final step of 10 min at 72 °C. Five microliters of the product were used for nested PCR, by using internal primers the internal primers, K80 5’AGCGTCTAGCCATGGCGT3’ and K78 5’CACTCGCAAGCACCCTATCAGGCAGT3’. The nested PCR mix consisted of 1 ×Pfu turbo buffer, 10 mmol/L of each dNTP, 20 pmol of internal primers, and 2.5 U of Pfu turbo DNA polymerase in a final volume of 100 μL. After a single denaturation step of 2 min at 95 °C, we carried out 30 cycles of 30 s at 95 °C, 30 s at 60 °C, and 2 min at 72 °C, and then, a final single step of 10 min at 72 °C. The amplified products of 240 nucleotides length were analyzed by electrophoresis onto 2% agarose gels stained with ethidium bromide. PCR products were purified by using the QIAquick PCR purification kit for direct sequencing on an Abi Prism 310 Genetic analyser (Applied Biosystems).
Extracted RNA was reverse transcribed using the degenerate primer NS5B8704 5’GADGAGCADGATGTWATBAGCTC3’ (nucleotide positions 8682-8704), where D = G + A + T, W = A + T and B = G + T + C, following the same conditions as for 5’UTR (see before). PCR was carried out by using the primer NS5B8256 5’TAYGAYACCMGNTGYTTTGACTC3’ (nucleotide positions 8256-8278), where Y = C + T, M = A + C, and N = A + T + G + C, with an initial denaturation step of 2 min at 95 °C, five initial cycles of 30 s at 95 °C, 30 s at 43 °C, and 2 min at 72 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 46 °C and 2 min at 72 °C, and completed with a single final step of 10 min at 72 °C. Heminested PCR was performed with an initial denaturation step of 2 min at 95 °C, 30 cycles of 30 s at 95 °C, 30 s at 48 °C, and 2 min at 72 °C, completed with a single final step of 10 min at 72 °C using primers NS5B8256 and NS5B8641 5’GARTAYCTGGTCATAGCNTCCGT3’ (nucleotide positions 8641-8619), where R = A + G, to obtain a final product of 386 nucleotides. Purification and sequencing were performed as mentioned above.
A GB query to Nucleotide collection (nr/nt), using the Megablast programme (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was performed for each of the NS5B sequences obtained in this study. The 100 GB sequences with the highest sequence similarity to each of our samples were selected. A tree was constructed by using either the Neighbor-joining or Fast Minimum Evolution algorithm from a given matrix of distances using the Jukes-Cantor method to calculate the distances. For every query, we obtained a tree that situates each sequence with the most closely related reference from the GB. The most similar sequences were downloaded for further analysis, the genotype was assigned, and a putative origin of local isolates was inferred. GB was accessed to download already published sequences of Argentine origin by searching with the words “HCV” and “Argentina” on the website: http://www.ncbi.nlm.nih.gov/sites/gquery. Published sequences obtained by several authors[15,17,22,23] were downloaded in Fasta format, and their length was then adjusted using the GeneDoc program[24].
Nucleotide sequences were resized by using GeneDoc and aligned by the CLUSTALW program[25]. Sequence similarity with other sequences from Argentina and with other GB sequences was ascertained by both distance and parsimony methods. Statistical support for each node in the trees drawn by both methods was obtained by performing 100 or 1000 bootstrap replicates of the original nucleotide sequence alignment. Phylogenetic analysis was carried out with the PHYLIP package[26]. Trees were drawn by using the Treeview program, v. 1.6.5[27].
Genetic divergences were calculated by using the DNASP program (version 3.53)[28] in three populations: our sequences grouped as a population, the closest GB sequences as another, and other Argentine sequences as the third one. Pairwise distances were calculated by using the MEGA3.1 program.
They were performed by using either the GraphPath Prism (version 5.0 for Windows) or the SigmaStat software. The non-paired Student’s t test was used to analyze the statistical differences between the mean age ± SD of the whole population of the country regarding those values recorded from each Province (therefore, examining a data set from two groups), as well as for such comparison with previous studies. When such study was performed among three or more groups, the one-way analysis of variance (ANOVA) was applied. Pairwise distances were statistically compared by using the χ2 test with Yates’ correction (SigmaStat software). P values lower than 0.05 were considered statistically significant.
A total of 6251 serum samples from a cohort of random volunteers was studied. Initially, 25 samples (0.40%) tested anti-HCV antibody positive as determined by EIA. However, 5 of the 25 samples failed to exhibit specific anti-HCV antibodies by immunoblotting analysis and also tested negative by RT-nested PCR to detect HCV RNA; hence, they were discarded for further studies.
HCV RNA was detected in serum samples from 7 out of 12 provinces. The prevalence of ongoing HCV infection as determined by RT-nested PCR from 12 of 23 provinces of Argentina, representing 73% of the country’s total population, was 0.32%. The highest prevalence was detected in Buenos Aires province (0.62%; 9/1461), a geographical area inhabited by 40.52% of the total population of Argentina. In the remaining provinces studied, the prevalence ranged from 0% in Chaco, Chubut, Entre Ríos, Jujuy, and San Luis, to up to 0.53% in Santiago del Estero. Intermediate values were observed in Salta (0.18%) and Córdoba (0.19%), Río Negro (0.30%), as well as in Catamarca (0.46%) and Tucumán (0.45%).
Genotype 1 was the most prevalent, accounting for 55% (11/20) of infected individuals, 25% were subtype 1a (5/20) and 25% subtype 1b (5/20). The second in prevalence was genotype 2 accounting for 35% (7/20); most sequences were ascribed to subtype 2c (n = 5), except one that clustered much closer to subtype 2j reference sequences. Lastly, genotype 3 accounted for the remaining 10% (2/20; Table 2). One genotype 1, one genotype 2 , as well as one genotype 3 isolates (according to their initial 5’UTR genotype assignment) were excluded from further analysis because of their respective NS5B RT-heminested PCR amplification failure.
| HCV genotype | HCV[+] 5’UTR sequences | Relative | From total population (n = 6251) | HCV subtype | HCV[+] NS5B sequences | Relative | From total population (n = 6251) |
| 1 | 11 | 55.00% | 0.176% | 1a | 5 | 25.00% | 0.080% |
| 1b | 5 | 25.00% | 0.080% | ||||
| 11 | 5.00% | 0.016% | |||||
| 2 | 7 | 35.00% | 0.112% | 2c | 5 | 25.00% | 0.080% |
| 2j | 1 | 5.00% | 0.016% | ||||
| 21 | 5.00% | 0.016% | |||||
| 3 | 2 | 10.00% | 0.032% | 3a | 1 | 5.00% | 0.016% |
| 31 | 5.00% | 0.016% | |||||
| Total | 20 | 100.00% | 0.320% | - | 17 | 100.00% | 0.320% |
NS5B phylogenetic trees and divergence analysis were performed by using the five HCV-1a, five HCV-1b, and five HCV-2c sequences. However, divergence could not be ruled out with those genotypes encompassing one single representative sequence (as occurred with 2j and 3a).
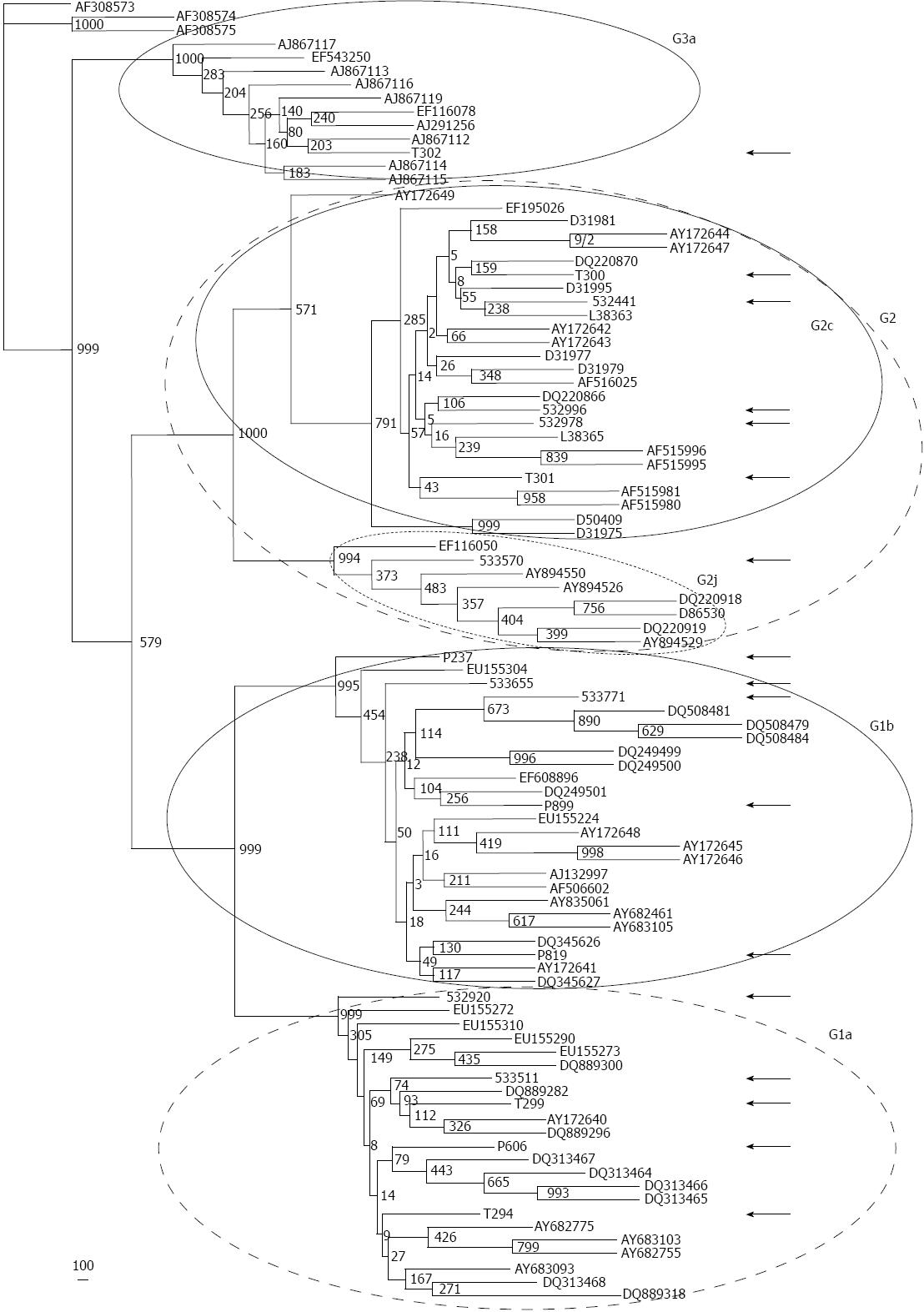
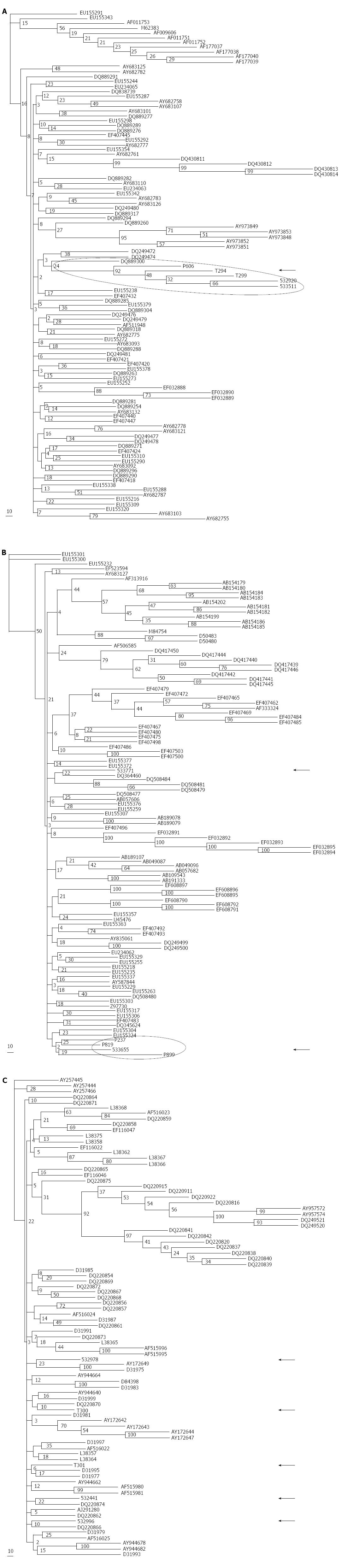
As expected, local NS5B sequences clustered together with their corresponding GB references (Figure 1), but the profile differed depending on the genotype. All of our HCV-1a Argentinian sequences clustered together with strong bootstrap support (92%; Figure 2A) and exhibited a low genetic divergence (0.034; Table 3). Furthermore, genetic divergence was not statistically different in comparison with other Argentinian sequences deposited in GB or with the closest non-Argentinian GB deposited sequences included in the study, suggesting a putative common source of infection/transmission.
| Argentine general population sequences | GB deposited sequences from Argentina | Closest GB deposited sequences | |
| HCV-1a | |||
| Argentine general population sequences | 0.034 | ||
| GB deposited sequences from Argentina | 0.042 | 0.040 | |
| Closest GB deposited sequences | 0.034 | 0.037 | 0.032 |
| HCV-1b | |||
| Argentine general population sequences | 0.082 | ||
| GB deposited sequences from Argentina | 0.074 | 0.046 | |
| Closest GB deposited sequences | 0.054 | 0.050 | 0.045 |
| HCV-2c | |||
| Argentine general population sequences | 0.087 | ||
| GenBank deposited sequences from Argentina | 0.092 | 0.098 | |
| Closest GB deposited sequences | 0.076 | 0.080 | 0.076 |
In the case of Argentinian HCV-1b, we observed two clusters with very low bootstrapping support (19%-25%), and one sequence distantly located regarding such clusters (Figure 2B). Genetic divergence was higher (0.082) than that observed among GB deposited Argentinian 1b sequences but the differences were not statistically significant. Similarly, HCV-2c sequences were intermingled along the tree with no particular clustering (Figure 2C) and showed a high genetic divergence (0.087). Phylogenetic analysis of Argentinian HCV-1b and HCV-2c sequences suggested a different origin of infection/transmission when compared with HCV-1a sequences.
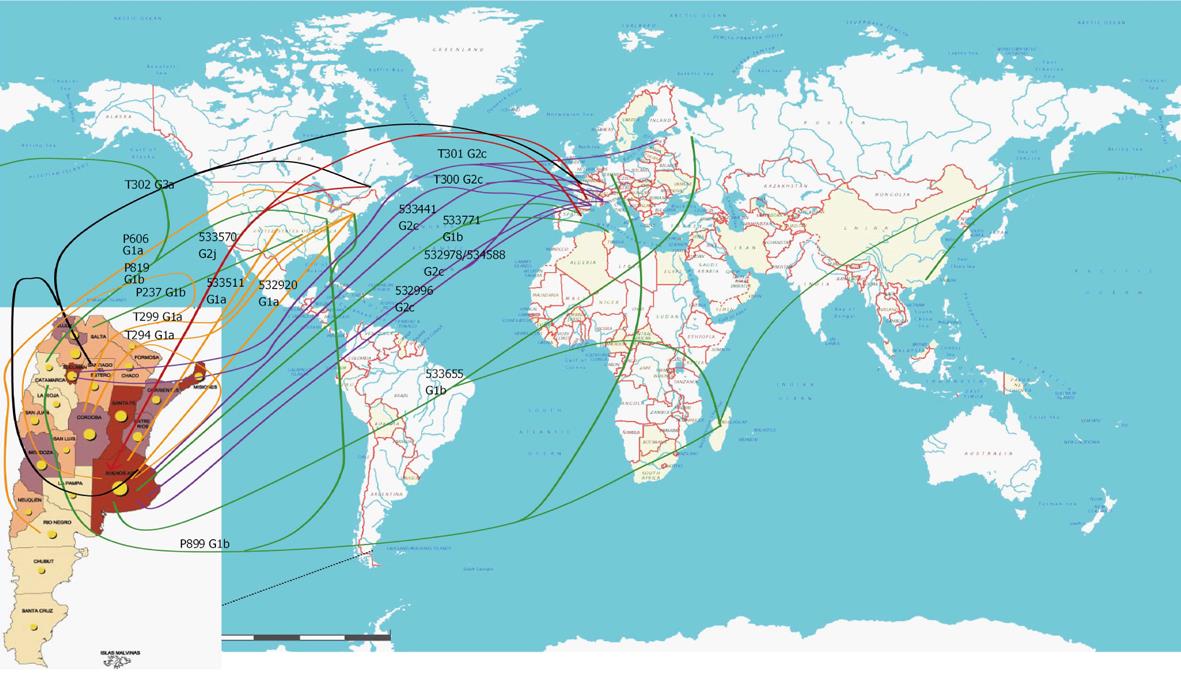
The closest GB sequences obtained by distance (UPGMA and NJ) and parsimony methods (DNAPARS) are represented in Table 4 (trees not shown). The geographic localization of the closest GB sequences is represented in Figure 3.
| Isolate | NS5B genotype | Closest GB deposited sequences | Origin |
| 532920 | 1a | DQ8893001bos | United States (Massachusetts, Boston area) |
| 533511 | 1a | EU155310usa EU155290usa AY683093usa DQ8893001usa | United States |
| P606 | 1a | AY682755alb AY683103alb AY682775alb DQ889318bos DQ889300bos | United States (Albany, NY) United States (Massachusetts, Boston area) |
| T294 | 1a | DQ313467arg DQ313464arg DQ313465arg DQ313466arg DQ889300bos | Argentina United States (Massachusetts, Boston area) |
| T299 | 1a | DQ889300bos DQ889296bos AY172640arg DQ889296bos | United States (Massachusetts, Boston area) Argentina United States (Massachusetts, Boston area) |
| 533655 | 1b | AF506602rus DQ345627mad | Russia (Western Siberia) Madagascar (Antananarivo) |
| 533771 | 1b | DQ508484tun DQ508481tun DQ508479tun EF608896bcn | Tunisia (Tunis) Spain (Barcelona) |
| P237 | 1b | EU155224ten EU155304ten | United States (Tennessee) |
| P819 | 1b | AY835061chn DQ249500usa DQ249499usa DQ345626mad | China (Foshan) United States (factor VIII concentrate) Madagascar (Antananarivo) |
| P899 | 1b | DQ345626mad AJ132997ger AY682461alb AY683105alb DQ249501usa | Madagascar (Antananarivo) Germany United States (Albany, NY) United States (factor VIII concentrate) |
| 532441 | 2c | L38363swi D31981ita | Switzerland Italy (France from Italians) |
| 532996 | 2c | L38365swi AF15995mar AF515996mar DQ220866tou | Switzerland France (Marseille) France (Toulouse) |
| 532978 534588 | 2c | D31975ita AY172649ita D50409(BEBE1)ita | Italy (France from Italians) |
| T300 | 2c | DQ220870fr | France (Toulouse) |
| T301 | 2c | AF516025mar EF195026tall AY944641gen D31977ita | France (Marseille) Estonia (Tallinn) Italy (Genoa) Italy (France from Italians) |
| 533570 | 2j | AY89526que AY894529que AY894550que EF116050que DQ220919tou DQ220918tou D86530bcn | Canada (Quebec) France (Toulouse) Spain (Barcelona) |
| T302 | 3a | EF116078que AJ291256ssd AJ867113arg AJ867159arg AJ867116arg AJ867115arg AJ867114arg | Canada (Quebec) France (Seine Saint Denis district) Argentina |
All local HCV-1a sequences grouped with GB United States sequences (St. Louis, Boston, or New York areas). The results suggest a narrow source of infection and not a multifocal event, and are consistent with the low degree of divergence found in the Argentine general population, despite the fact that the subjects studied resided in three different provinces (Buenos Aires, Córdoba, and Río Negro), and from whom serum sampling was performed, several hundred of kilometers apart from each other.
HCV-1b sequences grouped with those from all over the world, including Europe (Spain, Russia), Asia (China), Africa (Madagascar, Tunisia) and North America (United States), and represented a heterogeneous population (Table 3).
HCV-2c sequences from the Argentine general population formed a heterogeneous group with a completely different pattern as compared with HCV-1b. The sequences clustered with GB sequences originated from HCV patients in Italy. Even the sequences reported from Southeastern France were obtained from Italians living in this region. Only one of the clustered GB sequences was documented in Estonia. Three of our 2c sequences were detected in Buenos Aires/C.A.B.A. and two in Tucumán (approximately 1200 km from C.A.B.A.).
When blasted with GB sequences, the single sequence assigned to HCV-2j clustered with those from Canada, France, and Spain, showing the high heterogeneity among the HCV-2j sequences analyzed.
Regarding the single HCV-3a local sequence, similarities were found with one sequence from Canada and another one from France, and with an additional group of five sequences reported in Argentina.
Here we report a very low prevalence of HCV infection (0.32%) in a large cohort of random volunteers from Argentina, contrasting with the 2% prevalence previously reported in studies based on selected populations in small communities, or even higher rates among highly vulnerable groups[12,29] (Hepatitis C Argentinian Consensus, 2007). The observed prevalence is lower than that reported in neighboring countries, such as Brazil (1.5%), Uruguay (1%), and Chile (0.85%)[12]. The highest prevalence was detected in Buenos Aires province and C.A.B.A., both making up the region that received the greatest number of European immigrants, especially during the first half of the 20th century (70.38% of all immigrants, 20.8% residing in C.A.B.A.; http://www.mininterior.gov.ar/poblacion/estadisticas.asp Censo2001). In this regard, recent data shows that at present most of the European immigrants from Italy and Spain are over 60-year-old (http://www.mininterior.gov.ar/provincias/archivos_prv25/6-%20Perfil_Migratorio_de_la_Argentina.pdf).
The HCV isolates studied here did not form a close national cluster separate from the GB sequences. Interestingly, genetic divergence and phylogenetic analyses showed a different profile depending on the subtype analyzed. In this sense, the HCV-1a samples, detected from subjects residing in distantly placed cities/towns (hundreds of kilometers apart from each other) from three provinces, made up a highly homogeneous population, whereas the HCV-1b and HCV-2c samples were more heterogeneous, suggesting a different profile of epidemiological origin/transmission of infection for each subtype. The high homogeneity of subtype 1a and its similarity with sequences reported from United States suggest that HCV-1a was introduced in Argentina by a common infectious source from this geographic area. This finding agrees with the model of recent HCV genotype diversification in Central and South America[30-32] compared with other continents. HCV-1b isolates formed separate clusters that were most similar to European sequences, suggesting multiple focal transmission events, likely with independent geographical origins. Interestingly, the subject whose HCV isolate showed an HCV-1b phylogenetic relationship with a Russian HCV-1b sequence stated such ethnicity. HCV-2c represents an important contribution to Argentinian HCV epidemiology (at least, 25% in this study), supporting previous observations (23%)[15]. Most of the 2c isolates clustered close to sequences reported from Italy and Southern France. In general, the 2c sequences deposited in GB represent a highly heterogeneous population with huge genetic diversity in Ghana, Guinea, Benin, and Burkina Faso in Africa[33], suggesting that HCV-2c has long been present in human populations, especially in these parts of Africa, and that it spread to Egypt, Europe and elsewhere in the 20th century[34]. It has been proposed that the introduction of HCV-2c in Italy was a consequence of close contacts between native Africans and soldiers and colonials during the colonial wars in 1882-1896 and 1911-1912[35,36]. A high prevalence of HCV-2c was observed among individuals in Italy[37-40] and Southern France, all related with Italian immigrants[41]. Coincidentally, a substantial percentage of the Buenos Aires population descends from Italian immigrants that arrived in the 20th century. Taken together, our results suggest that the introduction of HCV-2c in Argentina may have been the result of a multiple event, likely related to waves of Italian immigration. In this regard, it is worth mentioning that a high prevalence (approximately 50%) of this genotype has been reported among chronic HCV patients from Córdoba province[19,20,29], as compared with data from Buenos Aires and C.A.B.A. patients[15] and even higher rates (90%) from patients residing in Cruz del Eje, a small rural town located in the Northern region of Córdoba province, where HCV prevalence was reported to be 5%[29]. In contrast, the present study could not detect the circulation of such genotype from the general population studied in the city of Córdoba (encompassing the whole group from the homonymous province). Several hypothetical factors might have contributed to the observed discrepancy, among them, it seems worth mentioning: (1) the previously reported overall low prevalence of infection in the city of Córdoba[29] and in this study; (2) the lower contribution of HCV-2c to the total HCV genotype prevalence in such capital city located in the central region of the province, as compared with Cruz del Eje[20,29]; (3) the dissimilar nature of studied groups (patients versus general population), hence showing a dissimilar HCV infection prevalence, and consequently having lower probability to pick up HCV positive samples; and (4) the mean age ± SE of all the analyzed populations (49.77 ± 2.15 for patients from Córdoba and other locations of Córdoba Province (n = 26)[29], 66.15 ± 1.52 years for patients from Cruz del Eje (n = 49)[29], as compared with 37.1 ± 0.4 in this study (SD = 12.5; median age = 35 years; n = 668). However, the last mentioned factor failed to reach statistical significance when the one way analysis of variance was carried out (P = 0.1177).
The recorded HCV-3a sequence exhibited similarities with isolates from France and Canada and other Argentinian isolates, in concordance with a more recent, worldwide expansion of this subtype[22]. The HCV-2j sequence showed similarities with French, Canadian, and Spanish HCV sequences. No other genotypes (4, 5 or 6) were detected in the Argentine general population studied.
In conclusion, NS5B analysis allowed an accurate classification of subtypes and enabled to perform the study of the evolution and origin of HCV infection. Here, we report a very low prevalence of HCV in the Argentine general population (0.32%). Phylogenetic analysis suggests diverse profiles of epidemiological expansion of HCV in Argentina: HCV-1a might have occurred from a putative common source, whereas HCV-1b and HCV-2c might have been introduced into the country following fluxes of immigration from other endemic areas, especially from Europe. The significantly high number of HCV-2c sequences compared to the reported data from neighboring countries may be the consequence of the high percentage of Italians migrating to Argentina from an area where such subtype was (and still is) highly prevalent. Argentina is a good example of how human practices, together with global expansion and human migration flows, have increased the HCV spread over the world. Adherence to standard universal precautions to avoid transmission should be strictly followed even in countries with a low prevalence of HCV.
We are indebted to Claudio Cenetrari, Darío Ciocale, Guillermo Colazo, Patricia Chenio, Maximiliano Gomes, Marina de los Santos, Luciana Novoa and the Fresenius Medical Care Centre for providing the blood samples. We thank both Celine Cavallo and Victoria Illas for English language support and helpful suggestions.
Hepatitis C virus (HCV) is a leading cause of chronic liver disease. HCV is distributed globally, affecting all countries with an estimated worldwide prevalence of 2.3% (approximately 160 million people) of the whole general population. Comparisons of HCV nucleotide sequences derived from individuals from different geographical regions revealed the circulation of at least six major HCV genotypes and more than 50 subtypes. Accurate HCV genotyping in chronically infected patients is crucial not only for epidemiological studies but also from a clinical standpoint, since the HCV genotype orientates the treatment strategy.
Direct sequencing, also referred as “population” sequencing, is the gold standard for HCV genomic sequence analysis. The viral genome region(s) sequenced must be carefully chosen, because not all of them provide accurate typing and subtyping. Since genotyping methods based on the exclusive analysis of the 5′NCR may lead to up to 10% mistyped results, there is a need to extend the analysis to coding regions such as NS5B or core. In this regard, the knowledge of the implicated HCV genotype in each patient contributes to select the appropriate treatment. Those infected with HCV genotype 1 must be treated with a triple combination of pegylated interferon-α (IFN-α), ribavirin and either telaprevir or boceprevir, whereas patients infected with other genotypes must still be treated with pegylated IFN-α and ribavirin alone. Moreover, HCV genotyping based on phylogenetic analysis, and - in case a representative sampling of a given (sub)genotype is obtained from an area - Monte Carlo Markov Chains Bayesian coalescent analysis may respectively lead to trace the origin and if such condition is met - the putative date of the Most Recent Common Ancestor of sequences.
This is a molecular epidemiological study performed in a large cohort of the local general population from 12 out of 23 Argentine provinces, as well as from the Autonomous city of Buenos Aires (the national capital). Unexpectedly, it shows a low prevalence of HCV (about 0.32%) in a general population cohort which included 6251 individuals. This low prevalence suggests that HCV could have been “recently” introduced in Argentina, as proposed by coalescent studies performed in restricted local areas of this country by other authors, where a predominant (sub)genotype was found, allowing such analysis. HCV subtypes were distributed as follows: 1a (25%), 1b (25%), 2c (25%), 3a (5%), and 2j (5%). HCV-1a sequences comprised a highly homogeneous population and clustered with United States sequences. HCV-1b sequences represented a heterogeneous population, suggesting that this genotype might have been introduced from different sources. Most HCV-2c sequences clustered close to the 2c reported from Italy and Southern France. Phylogenetic analysis is used by the authors to trace the putative source of HCV transmission and suggests that introduction of local HCV in this country is a consequence of multiple events that differed for each subtype studied. Diverse epidemiological patterns of HCV spread in Argentina might have occurred.
These new data could be useful to implement suitable measures regarding HCV surveillance by Argentine Public Health authorities.
HCV genotype: group of HCV variants assigned to a given genetic groups (1-6) which differs from others by 31%-33% at the nucleotide level. HCV subtype (sub-genotype): group of more closely related HCV variants assigned to a given genetic group which differs from others by 20%-25% at the nucleotide level (named with lower case letters: i.e., a, b, c, etc.).
This is a very well done and written molecular epidemiological study which considers the investigation of the prevalence of HCV infection and subtype frequencies among adults in Argentina. It should be underlined that authors have investigated a large amount of general population from 12 provinces representing all the geographical regions of the country.
P- Reviewers Vento S, Vorobjova T S- Editor Gou SX L- Editor A E- Editor Ma S
| 1. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1083] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 2. | Li G, Li K, Lea AS, Li NL, Abdulla NE, Eltorky MA, Ferguson MR. In situ hybridization for the detection of hepatitis C virus RNA in human liver tissue. J Viral Hepat. 2013;20:183-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [PubMed] |
| 4. | Domingo E, Holland JJ. High error rates, population equilibrium, and evolution of RNA replication systems in RNA Genetics. Boca Raton (FL): CRC Press 1988; 3-36. |
| 5. | Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 954] [Cited by in RCA: 831] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 6. | Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore EE, Peterson DA, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 263] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Quer J, Esteban JI, Cos J, Sauleda S, Ocaña L, Martell M, Otero T, Cubero M, Palou E, Murillo P. Effect of bottlenecking on evolution of the nonstructural protein 3 gene of hepatitis C virus during sexually transmitted acute resolving infection. J Virol. 2005;79:15131-15141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Hraber PT, Fischer W, Bruno WJ, Leitner T, Kuiken C. Comparative analysis of hepatitis C virus phylogenies from coding and non-coding regions: the 5’ untranslated region (UTR) fails to classify subtypes. Virol J. 2006;3:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, Poovorawan Y. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods. 2003;109:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Mellor J, Walsh EA, Prescott LE, Jarvis LM, Davidson F, Yap PL, Simmonds P. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J Clin Microbiol. 1996;34:417-423. [PubMed] |
| 11. | Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol. 2007;45:1102-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 215] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 12. | Kershenobich D, Razavi HA, Sánchez-Avila JF, Bessone F, Coelho HS, Dagher L, Gonçales FL, Quiroz JF, Rodriguez-Perez F, Rosado B. Trends and projections of hepatitis C virus epidemiology in Latin America. Liver Int. 2011;31 Suppl 2:18-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Golemba MD, Di Lello FA, Bessone F, Fay F, Benetti S, Jones LR, Campos RH. High prevalence of hepatitis C virus genotype 1b infection in a small town of Argentina. Phylogenetic and Bayesian coalescent analysis. PLoS One. 2010;5:e8751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Picchio GR, Baré PC, Descalzi VI, Bussy MV, Soria SM, Raffa MP, Mazzencio NE, Etchehun S, Cámera JA, Mosier DE. High prevalence of infection with a single hepatitis C virus genotype in a small rural community of Argentina. Liver Int. 2006;26:660-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Quarleri JF, Robertson BH, Mathet VL, Feld M, Espínola L, Requeijo MP, Mandó O, Carballal G, Oubiña JR. Genomic and phylogenetic analysis of hepatitis C virus isolates from Argentine patients: a six-year retrospective study. J Clin Microbiol. 2000;38:4560-4568. [PubMed] |
| 16. | Picchio GR, Nakatsuno M, Boggiano C, Sabbe R, Corti M, Daruich J, Pérez-Bianco R, Tezanos-Pinto M, Kokka R, Wilber J. Hepatitis C (HCV) genotype and viral titer distribution among Argentinean hemophilic patients in the presence or absence of human immunodeficiency virus (HIV) co-infection. J Med Virol. 1997;52:219-225. [PubMed] |
| 17. | Alfonso V, Flichman D, Sookoian S, Mbayed VA, Campos RH. Phylogenetic characterization of genotype 4 hepatitis C virus isolates from Argentina. J Clin Microbiol. 2001;39:1989-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Findor JA, Sordá JA, Daruich J, Bruch Igartua E, Manero E, Avagnina A, Benbassat D, Rey J, Nakatsuno M. [Distribution of the genotypes of hepatitis C virus in intravenous drug addicts in Argentina]. Medicina (B Aires). 1999;59:49-54. [PubMed] |
| 19. | Ruíz RD, Bísaro LB, German JC, Grutadauria S. [In Process Citation]. Rev Gastroenterol Mex. 2010;75:287-292. [PubMed] |
| 20. | Ré V, Lampe E, Yoshida CF, de Oliveira JM, Lewis-Ximénez L, Spinsanti L, Elbarcha O, Contigiani M. Hepatitis C virus genotypes in Córdoba, Argentina. Unexpected high prevalence of genotype 2. Medicina (B Aires). 2003;63:205-210. [PubMed] |
| 21. | Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2583] [Cited by in RCA: 2475] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 22. | Morice Y, Cantaloube JF, Beaucourt S, Barbotte L, De Gendt S, Goncales FL, Butterworth L, Cooksley G, Gish RG, Beaugrand M. Molecular epidemiology of hepatitis C virus subtype 3a in injecting drug users. J Med Virol. 2006;78:1296-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Gismondi MI, Becker PD, Valva P, Guzmán CA, Preciado MV. Phylogenetic analysis of previously nontypeable hepatitis C virus isolates from Argentina. J Clin Microbiol. 2006;44:2229-2232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Nicholas KB, Nicholas HB. GeneDoc: a tool for editing and annotating multiple sequence alignments. Available from: http://www.psc.edu/biomed/genedoc. |
| 25. | GCG , CLUSTALW . Wisconsin package version 8.1. Madison (WI): Genetic Computer Group 2002; . |
| 26. | Feltenstein J, PHYLIP , Distributed by the author. Department of Genetics, University of Washington, Seattle, United States. Available from: http: //evolution.genetics.washington.edu/phylip.html. |
| 27. | Roderic DM. TREEVIEW v1.6.5. Distributed by the authors. Available from: http: //taxonomy.zoology.gla.ac.uk/rod/treeview.html. |
| 28. | Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1496] [Cited by in RCA: 1385] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 29. | Forbi JC, Purdy MA, Campo DS, Vaughan G, Dimitrova ZE, Ganova-Raeva LM, Xia GL, Khudyakov YE. Epidemic history of hepatitis C virus infection in two remote communities in Nigeria, West Africa. J Gen Virol. 2012;93:1410-1421. [PubMed] |
| 30. | Ndjomou J, Pybus OG, Matz B. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J Gen Virol. 2003;84:2333-2341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693-712. [PubMed] |
| 32. | Cristina J. Genetic diversity and evolution of hepatitis C virus in the Latin American region. J Clin Virol. 2005;34 Suppl 2:S1-S7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Jeannel D, Fretz C, Traore Y, Kohdjo N, Bigot A, Pê Gamy E, Jourdan G, Kourouma K, Maertens G, Fumoux F. Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J Med Virol. 1998;55:92-97. [PubMed] |
| 34. | Pybus OG, Charleston MA, Gupta S, Rambaut A, Holmes EC, Harvey PH. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323-2325. [PubMed] |
| 35. | Ansaldi F, Bruzzone B, Salmaso S, Rota MC, Durando P, Gasparini R, Icardi G. Different seroprevalence and molecular epidemiology patterns of hepatitis C virus infection in Italy. J Med Virol. 2005;76:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 36. | Ndjomou J, Kupfer B, Kochan B, Zekeng L, Kaptue L, Matz B. Hepatitis C virus infection and genotypes among human immunodeficiency virus high-risk groups in Cameroon. J Med Virol. 2002;66:179-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Cammarota G, Maggi F, Vatteroni ML, Da Prato L, Barsanti L, Bendinelli M, Pistello M. Partial nucleotide sequencing of six subtype 2c hepatitis C viruses detected in Italy. J Clin Microbiol. 1995;33:2781-2784. [PubMed] |
| 38. | Guadagnino V, Stroffolini T, Rapicetta M, Costantino A, Kondili LA, Menniti-Ippolito F, Caroleo B, Costa C, Griffo G, Loiacono L. Prevalence, risk factors, and genotype distribution of hepatitis C virus infection in the general population: a community-based survey in southern Italy. Hepatology. 1997;26:1006-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 263] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 39. | Maggi F, Vatteroni ML, Fornai C, Morrica A, Giorgi M, Bendinelli M, Pistello M. Subtype 2c of hepatitis C virus is highly prevalent in Italy and is heterogeneous in the NS5A region. J Clin Microbiol. 1997;35:161-164. [PubMed] |
| 40. | Spada E, Ciccaglione AR, Dettori S, Chionne P, Kondili LA, Amoroso P, Guadagnino V, Greco M, Rapicetta M. Genotyping HCV isolates from Italy by type-specific PCR assay in the core region. Res Virol. 1998;149:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 41. | Nakao H, Okamoto H, Tokita H, Inoue T, Iizuka H, Pozzato G, Mishiro S. Full-length genomic sequence of a hepatitis C virus genotype 2c isolate (BEBE1) and the 2c-specific PCR primers. Arch Virol. 1996;141:701-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |