Published online Jun 28, 2013. doi: 10.3748/wjg.v19.i24.3770
Revised: February 14, 2013
Accepted: March 21, 2013
Published online: June 28, 2013
Processing time: 194 Days and 14 Hours
AIM: To investigate the biological function of 14-3-3σ protein and to look for proteins that interact with 14-3-3σ protein in colon cancer stem cells.
METHODS: Reverse transcription polymerase chain reaction was performed to amplify the 14-3-3σ gene from the mRNA of colon cancer stem cells. The gene was then cloned into the pGEM-T vector. After being sequenced, the target gene 14-3-3σ was cut from the pGEM-T vector and cloned into the pGBKT7 yeast expression plasmid. Then, the bait plasmid pGBKT7-14-3-3σ was transformed into the yeast strain AH109. After the expression of the pGBKT7-14-3-3σ fusion protein in the AH109 yeast strain was accomplished, a yeast two-hybrid screening assay was performed by mating AH109 with Y187 that contained a HeLa cDNA library plasmid. The interaction between the 14-3-3σ protein and the proteins obtained from positive colonies was further confirmed by repeating the yeast two-hybrid screen. After extracting and sequencing the plasmids from the positive colonies, we performed a bioinformatics analysis. A coimmunoprecipitation assay was performed to confirm the interaction between 14-3-3σ and the proteins obtained from the positive colonies. Finally, we constructed 14-3-3σ and potassium channel modulatory factor 1 (KCMF1) siRNA expression plasmids and transfected them into colon cancer stem cells.
RESULTS: The bait plasmid pGBKT7-14-3-3σ was constructed successfully, and the 14-3-3σ protein had no toxic or autonomous activation effect on the yeast. Nineteen true-positive colonies were selected and sequenced, and their full-length sequences were obtained. We searched for homologous DNA sequences for these sequences from GenBank. Among the positive colonies, four coding genes with known functions were obtained, including KCMF1, quinone oxidoreductase (NQO2), hydroxyisobutyrate dehydrogenase (HIBADH) and 14-3-3σ. For the subsequent coimmunoprecipitation assay, the plasmids PCDEF-Flag-14-3-3σ, PCDEF-Myc-KCMF1, PCDEF-Myc-NQO2 and PCDEF-Myc-HIBADH were successfully constructed, and the sequences were further confirmed by DNA sequencing. The Fugene 6 reagent was used to transfect the plasmids, and fluorescence-activated cell sorting analysis showed the transfection efficiency was 97.8% after 48 h. The HEK 293FT cells showed the stable expression of the PCDEF-Flag-14-3-3σ, PCDEF-Myc-KCMF1, PCDEF-Myc-NQO2 and PCDEF-Myc-HIBADH plasmids. After anti-Myc antibody immunoprecipitation with Myc-KCMF1, Myc-NQO2 and Myc-HIBADH from cell lysates, the presence of Flag-14-3-3σ protein in the immunoprecipitated complex was determined by western blot analysis. The knock-down expression of the 14-3-3σ and KCMF1 proteins significantly inhibited cell proliferation and colony formation of SW1116csc.
CONCLUSION: Genes of the proteins that interacted with 14-3-3σ were successfully screened from a HeLa cDNA library. KCMF1 and 14-3-3σ protein may affect the proliferation and colony formation of human colon cancer stem cells.
Core tip: Of the 14-3-3 proteins, tumor-suppressor activity has most clearly been defined for 14-3-3σ. In the study, we constructed 14-3-3σ bait gene and expressed as a fusion to the GAL4 DNA-binding domain successfully. Using the yeast two-hybrid system, we found novel binding proteins from the HeLa cDNA library which closely interact with 14-3-3σ. Our results also suggest that 14-3-3σ may interact with potassium channel modulatory factor 1 (KCMF1) protein. The knock-down expression of 14-3-3σ and KCMF1 proteins significantly inhibited proliferation and colony formation of SW1116csc cells. So, 14-3-3σ and other proteins may be involved in proliferation and colony formation of human colon cancer stem cells.
- Citation: Zou J, Mi L, Yu XF, Dong J. Interaction of 14-3-3σ with KCMF1 suppresses the proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol 2013; 19(24): 3770-3780
- URL: https://www.wjgnet.com/1007-9327/full/v19/i24/3770.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i24.3770
The 14-3-3 proteins are among the most abundant proteins within the cell, having been initially identified in 1967 as a family of acidic proteins within the mammalian brain. This family of highly conserved proteins consisting of seven isotypes in human cells (β, γ, ε, η, σ, τ, ξ) plays crucial roles in regulating multiple cellular processes, including the maintenance of cell cycle checkpoints and DNA repair, the prevention of apoptosis, the onset of cell differentiation and senescence, and the coordination of cell adhesion and motility. All 14-3-3 proteins bind to phosphoserine/phosphothreonine-containing peptide motifs corresponding to the sequences RSXpSXP or RXXXpSXP[1]. Many 14-3-3-binding proteins contain sequences that closely match these motifs, although a number of ligands bind to 14-3-3 in a phospho-independent manner using alternative sequences that do not closely resemble these motifs. Pozuelo Rubio et al[2] recently used 14-3-3-affinity chromatography and mass spectrometry to identify more than 200 14-3-3-binding ligands. All of these ligands lost their ability to bind to 14-3-3 upon dephosphorylation by the serine/threonine phosphatase PP2A in vitro. Some of the proteins bound to the 14-3-3-affinity column contained sequences that closely matched the optimal binding motifs, while others diverged significantly from the consensus sequences, despite their apparent phospho-specific binding.
Among the 14-3-3 proteins, 14-3-3σ is the isoform most directly linked to cancer[3]. There are several lines of evidence indicating that 14-3-3σ acts as a tumour suppressor gene and that its inactivation is crucial in tumorigenesis. The downregulation of 14-3-3σ by CpG methylation is detected in adenocarcinoma of the breast (96%), squamous cell carcinoma of the vulva (60%), lung cancer (83%), hepatocellular carcinoma (89%), ovarian carcinoma (60%), endometrioid endometrial adenocarcinoma (74%), gastric adenocarcinoma (43%), basal cell carcinoma (68.3%), squamous cell carcinoma of the bladder, neuroendocrine tumours (85%), and prostate cancer (45%)[4-10]. In our former proteomic study on human colon cancer stem cells, we found the expression of 14-3-3σ was obviously increased in colon cancer stem cells compared with differentiated cancer cells[11]. 14-3-3σ may be involved in the course of self-renewal, proliferation and differentiation of colon cancer stem cells.
To better understand the role of 14-3-3σ in the tumorigenesis, self-renewal, and differentiation of stem cells, we used the yeast two-hybrid system to find novel binding proteins that interact with 14-3-3σ. The bait gene, 14-3-3σ, was expressed as a fusion to the GAL4 DNA-binding domain, while the HeLa cDNA library was expressed as a fusion to the GAL4 activation domain. When the bait and library fusion proteins interact, the DNA-binding domain and activation domain are brought into proximity, thus activating the transcription of the reporter genes. Using this system, we found novel binding proteins from the HeLa cDNA library that closely interact with 14-3-3σ. This investigation provides functional clues for further exploration into novel cancer-related proteins for the treatment of colon cancer.
All yeast strains and plasmids for the yeast two-hybrid experiments were obtained from Clontech (Palo Alto, CA, United States) as components of the MATCHMAKER two-hybrid system 3. The Escherichia coli (E. coli) strain DH5α was used to clone every shuttle plasmid. The pGBKT7 DNA binding domain (DNA-BD) cloning plasmid, pGADT7 AD cloning plasmid, pGBKT7-53 control plasmid, pGADT7, pGBKT7-Lam control plasmid and pCL1 plasmid were obtained from Clontech Ltd. (K1612-1). The HeLa MATCHMAKER cDNA library was also obtained from Clontech Ltd.
Taq DNA polymerase, T4 DNA ligase, EcoRI and BglII restriction endonuclease were purchased from Takara Company, Japan. Lithium acetate, semi-sulphate adenine, acrylamide and N,N’-bis-acrylamide were purchased from Sigma Company United States. Tryptone and yeast extracts were purchased from the Oxoid Company, United States. X-α-gal and the culture media YPDA, SD/-Trp, SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, SD/-Trp/-Leu/-His/-Ade were purchased from Clontech, United States.
Human colon cancer stem cells (SW1116csc) were isolated and maintained in serum-free DMEM/F12 medium supplemented with human recombinant epidermal growth factor (20 μg/L; Invitrogen), human recombinant basic fibroblast growth factor (20 μg/L; Invitrogen), L-glutamine (2 mmol/L), insulin (4 U/L), penicillin G (1 × 105 U/L), and streptomycin (100 mg/L). Total RNA was isolated with TRIzol reagent (Invitrogen, United States) according to the manufacturer’s instructions. The total RNA recovered from the DNase I digestion was measured at 260 and 280 nm with a spectrophotometer (Ultraspec 2000, Pharmacia Biotech), with the 260 nm reading used to estimate the concentration of total RNA. The 260/280-nm ratios and a 1% agarose-formaldehyde gel stained with ethidium bromide were used to confirm the RNA quality of the samples.
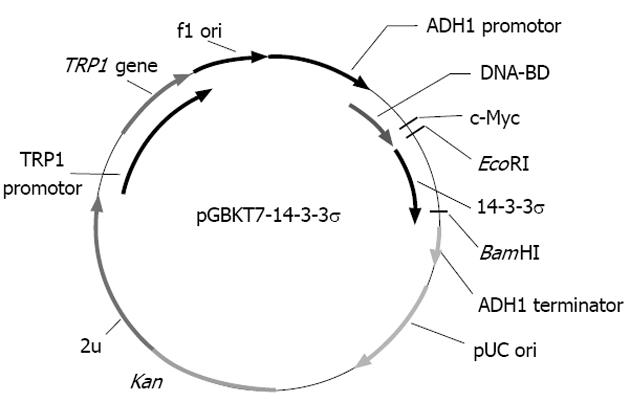
To make the bait plasmid, reverse transcription polymerase chain reaction (RT-PCR) was performed to amplify the 14-3-3σ gene from SW1116csc cells. The sequences of the primers contained EcoRI and BglII restriction enzyme sites. The PCR conditions were as follows: 94 °C for 45 s, 60 °C for 45 s, 72 °C for 1 min, for 35 cycles. Ten nanograms of the 747 bp PCR product were cloned into the pGEM-T vector. The primary structure of the insert was confirmed by direct sequencing. The fragment encoding 14-3-3σ was released from the pGEM-T-14-3-3σ by digestion with EcoRI and BglII and was then ligated into pGBKT7. Vector pGBKT7-expressing proteins were fused with amino acids 1-147 of the GAL4 DNA-BD, and pGADT7-expressing proteins were fused with amino acids 768-881 of the GAL4 activation domain. The plasmid pGBKT7-14-3-3σ, containing the full-length 14-3-3σ gene, could directly express the DNA binding domain, c-Myc and 14-3-3σ fusion protein. The plasmid was transformed into the yeast strain AH109 with the lithium acetate method.
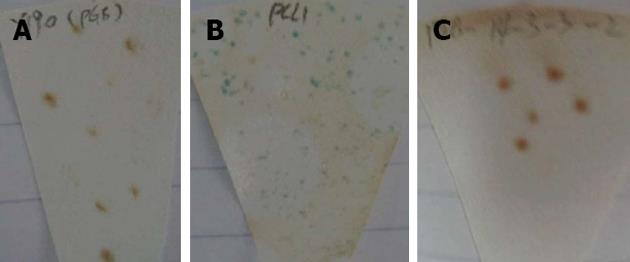
The purified bait plasmid was transformed into the AH109 strain and was then cultured on SD/-Trp/agar plates for detection. Approximately 2 mm of AH109 colonies, transformed by pGBKT7-14-3-3σ and pGBKT7, were incubated in 3 mLYPDA liquid medium at 30 °C for 16 h with shaking. The absorbance values at 600 nm (A600) in different groups were compared. Additionally, transformants containing the pGBKT7-14-3-3σ and pGBKT7 plasmids were transferred to SD/-Trp/X-a-gal, SD/-Trp/-his/X-a-gal and SD/-Ade/-Trp/X-a-gal plates at 30 °C for 5 d. In parallel, AH109 cells transformed by pCL1 and pGBKT7-Lam served as the positive and negative controls, respectively.
The screening of the HeLa cDNA library was performed as follows. One large (2-3 mm), fresh (< 2 mo old) colony of AH109 (bait) was inoculated into 50 mL SD/-Trp and incubated and shaken at 250-270 r/min at 30 °C overnight (16-24 h). Then, the cells were pelleted by centrifuging the entire 50-mL culture at 1000 r/min for 5 min. After the supernatant was decanted, the cell pellet was resuspended in the residual liquid by vortexing. A HeLa cDNA library was cloned into pACT2 and the yeast reporter strain Y187. The entire AH109 (bait) culture and 1 mL of the HeLa cDNA library were combined and cultured in a 2-L sterile flask; 45 mL of 2 × YPDA/Kan was added and swirled gently. After a 20-h mating period, the cells were pelleted, re-suspended and spread on 50 large (150 mm) plates containing 100 mL SD/-Ade/-His/-Leu/-Trp. After 6-15 d, the yeast colonies were transferred onto plates containing X-α-gal to evaluate the expression of the MEL1 reporter gene (blue colonies).
Approximately 1 × 106 colonies were screened, and positive clones were identified. The yeast plasmids were isolated from the positive colonies with the lyticase method and transformed into super-competent E. coli DH5α by electroporation. The transformants were plated on ampicillin SOB selection media and grown under selection conditions. Subsequently, the pACT2-cDNA constructs were re-isolated following the standard protocol, analysed by restriction digestion and sequenced.
After the positive colonies were sequenced, the sequences were blasted against sequences in GenBank to analyse the function of the genes (http://www.ncbi.nlm.nih.gov/blast). Other bioinformatics analyses, including their molecular weight, theoretical PI, estimated half-life, secondary structure prediction, and so on, were respectively performed with different software tools (http://www.expasy.ch/tools/protparam.html; http://www.cmpharm.ucsf.edu/nomi/ nnpredict.html; http://www.ch.embnet.org/software/COILS_form.html; http://www.expasy.org/tools/protscale. html; http://www.ch.embnet.org/software/TMPRED_ form.html; http://www.cbs.dtu.dk/services/SignalP/).
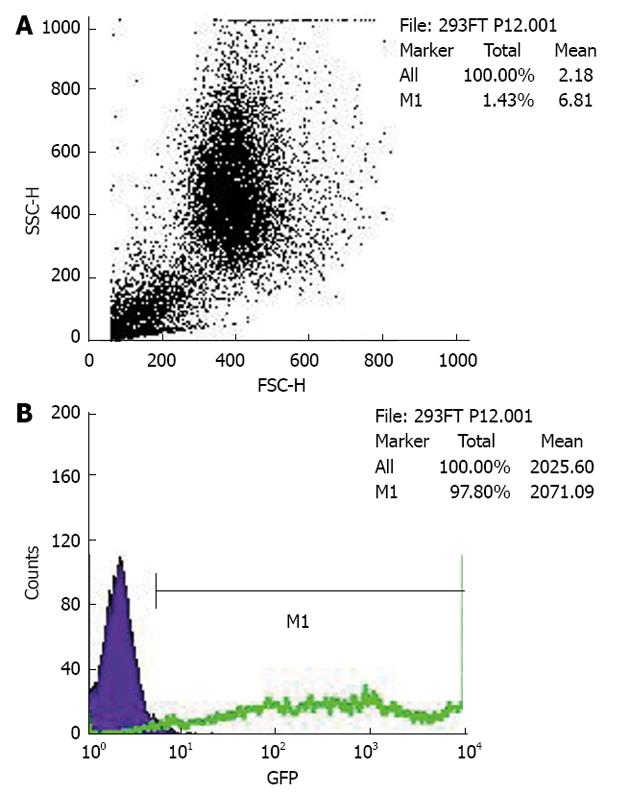
The plasmids in positive yeast clones were isolated from the colonies by the lyticase method. The 14-3-3σ, potassium channel modulatory factor 1 (KCMF1), quinone oxidoreductase (NQO2) and hydroxyisobutyrate dehydrogenase (HIBADH) genes were PCR amplified with specific primers, and the products were characterised by restriction digest using SfiI. After PCDEF-Flag and PCDEF-Myc plasmids were digested with the SfiI restriction enzyme, 14-3-3σ, KCMF1, NQO2 and HIBADH cDNA were cloned into them. The correct plasmids were named PCDEF-Flag-14-3-3σ, PCDEF-Myc-KCMF1, PCDEF-Myc-NQO2 and PCDEF-Myc-HIBADH. The plasmid sequences were analysed by DNA sequencing. HEK 293FT cells (5 × 105 cells) were cultured in a 6-cm dish until 60% confluence was reached. Various types of plasmids were transfected using the Fugene 6 Transfection Kit (Roche) according to the manufacturer’s instructions (Table 1). Briefly, 1.5 μL Fugene 6 was diluted with 100 μL Opti-MEMI (Invitrogen), mixed, and incubated at room temperature for 5 min. A 0.5 μg quantity of plasmid was added to the Fugene 6/Opti-MEM combination at a 3 (DNA):1 (Fugene) ratio, then mixed and incubated at room temperature for 45 min. The mixture of Fugene 6 and plasmid DNA was then added to cells cultured in 0.5 mL fresh Opti-MEM and incubated at 37 °C in 5% CO2. After 14-16 h, the transfection reagent was replaced with fresh culture medium; 48 h later, the transfected cells were harvested and fluorescence-activated cell sorting (FACS) was performed.
| Plasmid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Transfection control | |
| Sample 1 | Negative | Sample 2 | Negative | Sample 3 | Negative | Negative | Positive | Positive | Negative | |
| KCMF1 | Control-1 | NQO2 | Control-2 | HIBADH | Control-3 | Control-4 | ||||
| Flag-mfrP53 | - | - | - | - | - | - | - | + | pEGFP | 293FT |
| Myc-Large T | - | - | - | - | - | - | - | + | ||
| Flag-14-3-3 | + | - | + | - | + | - | + | - | ||
| Myc-HIBADH | - | - | - | - | + | + | - | - | ||
| Myc-NQO2 | - | - | + | + | - | - | - | - | ||
| Myc-KCMF1 | + | + | - | - | - | - | - | - | ||
| VECTOR-Myc | - | - | - | - | - | - | - | - | ||
| VECTOR-Flag | - | + | - | + | - | + | - | - | ||
FACS of enhanced green fluorescent protein (EGFP) positive cells was performed on a FACS Aria (Becton Dickinson). The cells were washed twice with Ca2+- and Mg2+-free HBSS and then incubated with 50 U papain (Worthington) and 100 U DNase I (Sigma) in PIPES at 37 °C for 10 min with gentle shaking. The samples were spun, resuspended in 2 mL DMEM/F12/N2 and dissociated by sequentially titrating with three serially narrowed glass Pasteur pipettes. The papain was inactivated with DMEM/F12/N2 plus 20% FBS, and the cells were pelleted. The pelleted cells were resuspended in HBSS and then passed over a 40 μm cell strainer. The cells were resuspended at a final concentration of (3-6) × 106 cells/mL with 1.0 μg 7-amino-actinomycin D (7-AAD). The cells were analysed by forward and side scatter for EGFP fluorescence through a 530 ± 30 nm bandpass filter and for 7-AAD fluorescence through a 695 ± 40 nm bandpass filter. The EGFP-positive cells were sorted at 2000-5000 events/s with a purification mode algorithm. Untransfected cells were used as a control to set the background fluorescence; a false-positive rate of 0.1% was accepted to ensure an adequate yield.
The HEK 293FT cell line was grown in accordance with the ATCC recommendations. Forty-eight hours after plasmid transfection, the cells were lysed in ice-cold 1% Triton X-100 buffer containing a cocktail of protease inhibitors. The lysates were cleared by centrifugation at 12000 g for 15 min at 4 °C. Coimmunoprecipitation assays using cleared cell lysates were performed at 4 °C for 2 h with the appropriate antibody. Immune complexes were precipitated with protein G Sepharose beads for an additional 1 h, washed three times with cold lysis buffer, resuspended in 16 Laemmli sample buffer, boiled for 5 min, subjected to SDS-PAGE and transferred to NC filters. The NC filters were blocked for 1 h at 4 °C in 5% nonfat milk in TBS (50 mmol/L Tris, 150 mmol/L NaCl) containing 0.1% Tween-20 (Sigma). They were then incubated for 2 h with primary antibodies (1:1000 dilution) in the blocking solution. After extensive washes in TBS 0.1% Tween-20, the filters were incubated for 1 h with HRP-conjugated anti-mouse antibody (Serotech) diluted 1:5000 in TBS 5% nonfat milk solution. After final washes in TBS 0.1% Tween, Western blottings were developed with the ECL kit from Amersham Biosciences.
Selection of the siRNA sequence was based on the siRNA Target Finder and Design Tool available at the Ambion Inc. web site and related reference. The siRNAs targeting human 14-3-3σ and KCMF1 mRNA common sequence 5’-CCCAGAAGAUGGACUUCUA-3’ and 5’-CGCGUGUCGAAGACUAUUU-3’ were synthesised and purified by Shanghai Sangon Corporation. The sense strand of the pU-siRNA inserts was 5’-GATCCACCTCACCAAGGCCAGCACTTCAAGAGAGCTGGCCTTGGTGAGGTTTTTTTTGGAAGTCGACA-3’; it was inserted into BamHI-HindIII linearised pRNAT-U6.1/neo vector (Ambion Inc., Austin, TX, United States). The inserted sequences were verified by DNA sequencing. The control siRNA vector (pU-siCONT; Ambion Inc.) was constructed by the insertion of a sequence that expressed a hairpin siRNA with no significant homology to any known sequences in the human, mouse or rat genomes. The control insert sequence was: 5’-GATCCACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTTGGAAA-3’. One microgram per well of pU-14-3-3σ-siRNA, pU-KCMF1-siRNA expression plasmid or control plasmid (pU-siCONT) were transfected into SW1116csc. Forty-eight hours later, transfection-positive cells were observed under a fluorescence microscope.
SW1116csc and siRNA-transfected SW1116csc cells were seeded at a density of 1 × 104 in 35-mm Petri dishes. The cultured cells stained with trypan blue were observed and counted in triplicate over 6 wk. The cells were disassociated, suspended in medium containing 0.3% agar, and plated onto a bottom layer containing 0.6% agar. The cells were plated at a density of 3 × 104 cells/6-cm dish, and the number of colonies that were > 0.5 mm in diameter was counted 14 d later.
Total RNA was extracted from 1 × 108 SW1116csc cells and yielded approximately 10 μg of high purity total RNA. The absorbance ratios of the RNA at 260/280 and 230/260 nm were 2.03 and 2.00, respectively, indicating that the RNA was of the highest quality and was therefore useful for the following experiments.
The bait plasmid pGBKT7-14-3-3σ was constructed with complete E6 cDNA successfully, and the results of sequencing analysis suggested that the cDNA was in-frame, no artefacts were added to the E6 sequence and the restriction sites were correct (Figure 1).
Two to three millimetre AH109 colonies, transformed by pGBKT7-14-3-3σ and pGBKT7, were incubated in 3 mL YPDA liquid medium. The A600 nm values in the AH109-pGBKT7-14-3-3σ and AH109-pGBKT7 groups were 0.98 and 0.99, respectively, which suggested that the pGBKT7-14-3-3σ plasmid was not toxic to yeast and had no effect on the growth of the yeast. Furthermore, the AH109-pGBKT7-14-3-3σ clones were white and were detected on the SD/-Trp/-His/X-α-gal and SD/-Ade/-Trp/X-α-gal plates (Figure 2). Therefore, the 14-3-3σ protein was believed to have no autonomous activation effect.
Diploids were detected under an inverted microscope 20 h after co-incubation, which indicated that yeast mating was successful. Nineteen positive colonies grew on the SD/-Ade/-His/-Leu/-Trp/X-α-gal agar medium and restreaked three times. Finally, 19 putative positive yeast colonies were obtained.
Nineteen positive yeast colonies were selected, and each purified plasmid DNA was directly transferred to competent DH5α cells by electroporation. The transformants containing only the pGADT7-HeLa cDNA library plasmids were obtained by plating on LB/Amp agar medium. Sequencing analysis was performed, and 4 genes that interacted with 14-3-3σ, including KCMF1, NQO2, HIBADH and 14-3-3σ, were identified (Table 2).
| Bait | Prey library | Identical colonies | Positive clone name | Positive gene identified | NCBI accession number | Gene code match | |
| pGB-14-3-3 | 19 | Sigma 12 | SFN | NM_006142.3 | Yes | Full | |
| Sigma 13 | Lengh | ||||||
| Sigma 30 | |||||||
| Human HeLa | Sigma 4 | HIBADH | NM_152740.3 | Yes | Full | ||
| MATCHMAKER | Sigma 15 | Lengh | |||||
| cDNA library | Sigma 21 | NQO2 | NM_000904.3 | Yes | Full | ||
| Sigma 32 | Lengh | ||||||
| Sigma 23 | |||||||
| Sigma 33 | Full | ||||||
| Sigma 28 | KCMF1 | NM_020122.4 | Yes | Lengh |
The diploids were detected under an inverted microscope, and blue colonies were grown on the SD/-Ade/-His/-Leu/-Trp/X-α-gal agar medium.
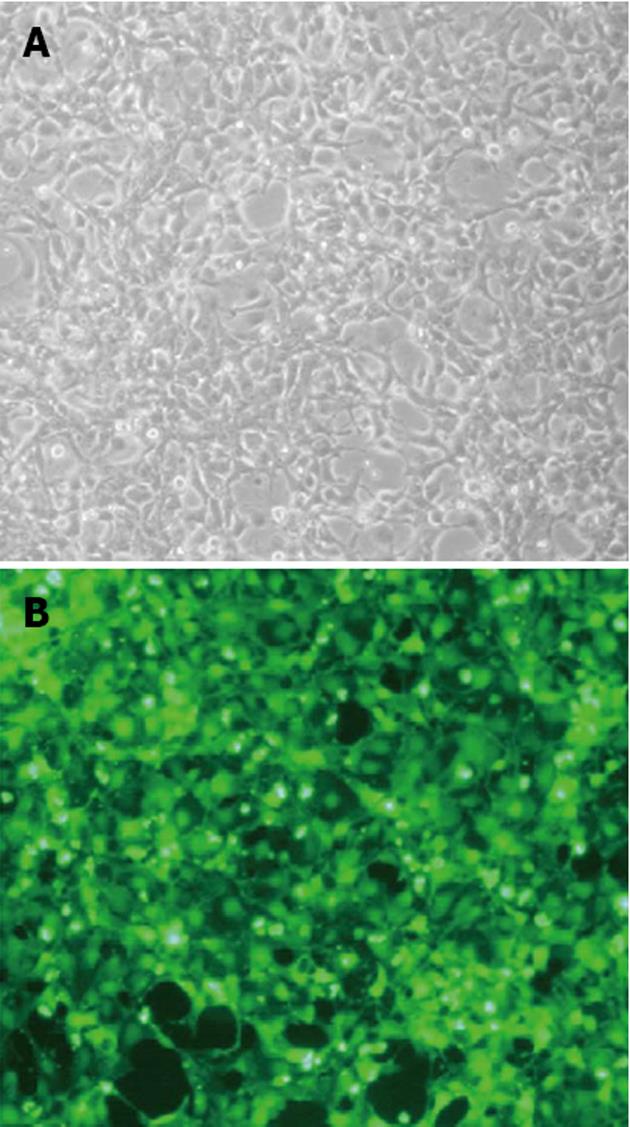
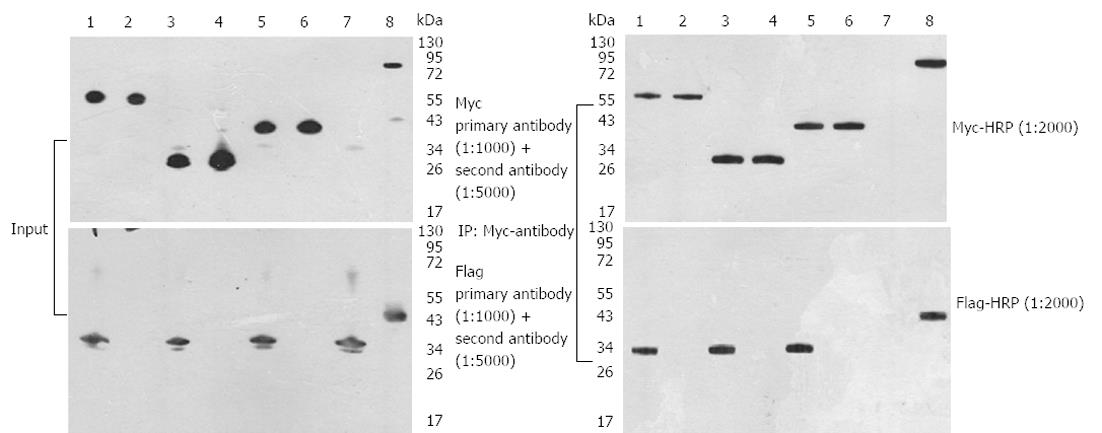
The PCDEF-Flag-14-3-3σ, PCDEF-Myc-KCMF1, PCDEF-Myc-NQO2 and PCDEF-Myc-HIBADH plasmids were successfully constructed. The sequences of the plasmids were further confirmed by DNA sequencing. The plasmids were transfected into HEK 293FT cells with the Fugene 6 reagent (Figure 3). After 48 h, FACS assay was performed and showed that the transfection efficiency was 97.8% (Figure 4).
Three interactions detected in this two-hybrid screen were further confirmed in HEK 293FT cells by specific coimmunoprecipitation of Flag-tagged bait proteins with the three Myc-tagged prey proteins. HEK 293FT cells stably expressed the PCDEF-Flag-14-3-3σ, PCDEF-Myc-KCMF1, PCDEF-Myc-NQO2 and PCDEF-Myc-HIBADH plasmids (Figure 5A). An anti-Myc antibody was used to immunoprecipitate Myc-KCMF1, Myc-NQO2 and Myc-HIBADH from cell lysates. The presence of Flag-14-3-3σ protein in the immunoprecipitated complex was determined by western blot analysis (Figure 5B). These coimmunoprecipitations confirm that several of the novel interactions identified in the present two-hybrid screen are reproducible in the context of mammalian cells and therefore validate the results obtained by the two-hybrid assay.
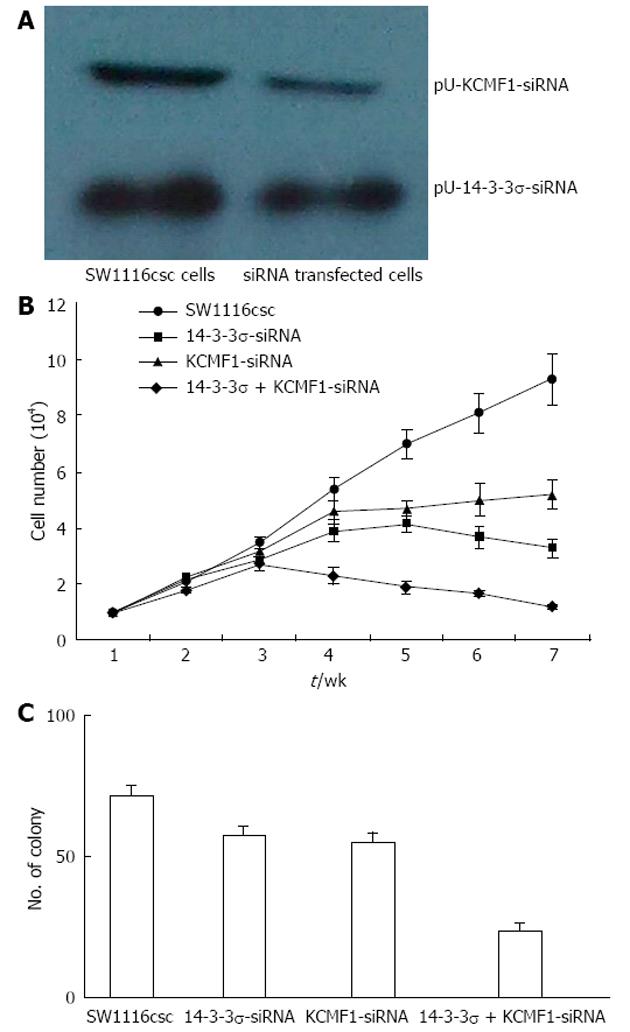
After pU-14-3-3σ-siRNA or pU-KCMF1-siRNA expression plasmid transfection, the knock-down of the 14-3-3σ and KCMF1 proteins in SW1116csc cells was observed (Figure 6A). We tested for differences in the proliferation rate between SW1116csc and siRNA-transfected SW1116csc. The cells were examined from week 1 to week 7 after seeding. As shown in Figure 6B, there was a difference in the growth rate between the transfected and control cells. The transfected cells grew slowly and showed growth inhibition after week 4. The self-renewing capacity of the transfected cells was also examined with the colony formation assay. When plated at a density of 100 cells/well, 14-3-3σ-siRNA, KCMF1-siRNA and 14-3-3σ + KCMF1-siRNA transfected cells generated a lower mean number of tumour spheres (57.4 ± 3.6, 55.3 ± 3.3 and 23.7 ± 2.8, respectively) compared to the SW1116csc cells (71.4 ± 4.1) (Figure 6C).
Protein-protein interactions occur in a wide variety of biological processes and essentially control cell fate from division to death. Yeast two-hybrid assays represent a versatile tool to study protein interactions in vivo. The yeast two-hybrid system 3, based on the system originally designed by Fields and Song, takes advantage of the properties of the GAL4 protein of the yeast Saccharomyces cerevisiae. The GAL4-based assay uses the yeast transcription factor GAL4 for the detection of protein interactions by transcriptional activation. GAL4 possesses a characteristic phenomenon in which the transactivation function can be restored when the factor’s DNA-binding domain (DBD) and its transcription-AD are brought together by two interacting heterologous proteins. The GAL4-yeast two-hybrid assay uses two expression vectors, one with DBD and the other with AD. The GAL4-DBD fuses to protein “X” and GAL4-AD fuses to protein “Y” to form the bait and the target of the interaction trap, respectively. A selection of host cells with different reporter genes and different growth selection markers provides a means to detect and confirm protein-protein interactions and has significantly fewer false positives.
To investigate the role of 14-3-3σ in tumorigenesis, the yeast two-hybrid system 3 was used to screen the proteins interacting with 14-3-3σ. In this study, the bait plasmid pGBKT7-14-3-3σ was transformed into the yeast strain AH109. To further confirm the expression of 14-3-3σ protein in the AH109 yeast strain, we performed Western blotting analysis and observed strong 14-3-3σ expression. After the bait plasmid pGBKT7-14-3-3σ yeast strain AH109 was mated with the HeLa cDNA library yeast strain Y187, resulting diploid yeast cells were plated on QDO media containing X-a-gal. Nineteen true positives were confirmed and obtained. Through sequencing analysis of isolated library plasmids, we obtained the sequences of four types of genes with known functions.
In addition to their well-known pro-proliferative and anti-apoptotic effects, 14-3-3 proteins have also been found to suppress cell growth and cell-cycle progression, especially after DNA damage, indicating functions in tumour suppression[12-15]. Of the 14-3-3 proteins, the tumour-suppressor activity has most clearly been defined for 14-3-3σ[16-19]. 14-3-3σ is unique among the 14-3-3 proteins in that it is expressed primarily in epithelial cells and forms homodimers almost exclusively[20]. Further insight as to why the loss of 14-3-3σ might facilitate tumor formation comes from the discoveries by Wilker et al[21] reported that 14-3-3σ is a crucial regulator of translation during mitosis and that 14-3-3σ function is required for proper mitotic exit and cytokinesis. In eukaryotic cells, most mRNA translation occurs via a cap-dependent mechanism in which ribosome recruitment begins with the binding of eukaryotic initiation factors, such as eIF4B, to a modified guanosine residue (known as a “cap’’) at the 5’ end of the mRNA. However, some mRNAs contain internal ribosome entry sites and are translated in a cap-independent manner. During mitosis, cap-dependent translation is suppressed and cap-independent translation is stimulated, allowing for the translation of key cell-cycle regulators such as cell division cycle 2-like 1. Experiments by Wilker et al[21] showed that 14-3-3σ is needed for the mitotic switch from cap-dependent to cap-independent translation and that 14-3-3σ appears to mediate this switch by binding to eIF4B and perhaps other factors involved in cap-dependent translation. When cells are depleted of 14-3-3σ, cap-dependent translation is not suppressed and cytokinesis is impaired, resulting in the generation of binucleated cells, a phenotype observed in the early stages of tumour formation.
14-3-3 acts as an adaptor or “chaperone molecule”, which is able to move freely from the cytoplasm to the nucleus and vice-versa[22]. 14-3-3 proteins are mainly cytoplasmic molecules; they can form homodimers or heterodimers, and interact with various cellular proteins. 14-3-3 proteins are phosphoserine-binding proteins that bind the consensus motifs RSXpSXP and RXY/FXpSXP. These consensus motifs are present in almost all of the 14-3-3 binding proteins[1]. More than a hundred small molecules interact with 14-3-3 in a phosphorylation-dependent manner. These proteins include protein kinases (murine leukaemia viral oncogene homologue-RAF1, MEK kinase, PI3 kinase and Grb10), receptor proteins (insulin-like growth factor 1 and glucocorticoid receptors), enzymes (serotonin N-acetyltransferase, tyrosine and tryptophan hydroxylase), structural and cytoskeletal proteins (vimentins and keratins), scaffolding molecules (calmodulin), proteins involved in cell cycle control (cdc25, p53, p27 and wee1) proteins involved in transcriptional control (histone acetyltransferase, and TATA box binding proteins), and proteins involved in apoptosis (BAD)[1,23]. However, a few proteins interact with 14-3-3 in a phosphorylation-independent manner such as Bax. Recently, using direct proteomic analysis, researchers have identified a large number of polypeptides (> 200) that can associate with 14-3-3 proteins. These polypeptides are involved in numerous cell functions, including fatty acid synthesis, reductive metabolism, iron and other metabolisms, DNA/chromatin interactions including transcription factors, RNA binding, protein synthesis, protein folding and processing, proteolysis, protease inhibitors, ubiquitin metabolism, cellular signaling and apoptosis, actin dynamics, cellular trafficking and transporters, signaling kinases, cell division, nuclear proteins, oncogenic signaling, and cytoskeletal proteins[2,24]. A study demonstrated that some of the 14-3-3 binding proteins are involved in the regulation of the cytoskeleton, GTPase functions, membrane signaling, and cell fate determination[25]. In this study, we found that 14-3-3σ could interact with the proteins KCMF1, NQO2, HIBADH and 14-3-3σ.
The function of NQO2 is not clearly understood. NQO2 is expressed selectively in the kidneys, skeletal muscles, liver, heart, and lungs, suggesting a tissue-specific action of the enzyme. Some studies have suggested that greater NQO2 expression may activate certain types of chemicals in the brain, leading to oxidative stress and neuronal damage[26]. Other studies have implied that NQO2 can protect against quinone-induced skin carcinogenesis[27]. Recently, new evidence has shown for the first time that NQO2 catalyses the reduction of electrophilic oestrogen quinones and thereby acts as a detoxification enzyme. Gaikwad et al[28] successfully demonstrated that oestrogen-3,4-quinone binds to NQO2 and established that oestrogen quinines are endogenous biological substrates of NQO2. Moreover, they demonstrated that NQO2 is faster at reducing oestrogen quinones than its homologue NQO1. Such findings reveal a possible relationship between breast cancer and NQO2, although no studies to date have addressed this issue. In addition, NQO2 can stabilise the p53 protein[29], a known breast tumor suppressor gene product. p53 is recognised as a highly penetrant breast cancer susceptibility gene, and loss of both p53 and breast cancer type 1 susceptibility protein (BRCA1) results in the rapid and efficient formation of mammary carcinomas[30]. Interestingly, the expression of 14-3-3σ is coordinately upregulated by the cellular tumour antigen p53 and BRCA1 and contributes to the DNA-damage-induced cell-cycle checkpoint mediated by these tumour suppressors[31]. It is logical to assume that 14-3-3σ binds to and sequesters NQO2 in the cytoplasm, thus enabling DNA damage to be repaired before the cell cycle progresses. In this study, we found that 14-3-3σ could interact with NQO2 directly and further confirmed the important function of 14-3-3σ protein in DNA repair and cell cycle progress.
KCMF1 encodes a zinc-finger protein with hitherto barely characterised function. KCMF1 was mentioned in 2001 at NCBI as an expressed sequence tag clone potentially involved in the regulation of potassium channels. A partial expressed sequence tag sequence of KCMF1 was identified as a differentially regulated gene during kidney tubulogenesis in vitro and designated as developing branching tubulogenesis 91 (Debt91)[32]. In addition, KCMF1 was shown to be downregulated in Ewing’s sarcoma cell lines after the overexpression of CD99 and upregulated through fibroblast growth factor (FGF) receptor signalling pathways in gastric cancer cells and was consequently named basic FGF induced in gastric cancer[33]. Kreppel et al[34] showed that the nuclear zinc-finger protein KCMF1 was overexpressed in epithelial cancers and especially in human and mouse pancreatic cancer. KCMF1 enhanced proliferation, migration and invasion. The downregulation of KCMF1 in vivo reduced preneoplastic changes in the transforming growth factor-α transgenic pancreatic cancer model. One study showed that 14-3-3σ was highly expressed in pancreatic adenocarcinoma[35]. Our results suggest that 14-3-3σ may also interact with KCMF1. The knock-down of 14-3-3σ and KCMF1 proteins expression significantly inhibited cell proliferation and colony formation of human colon cancer stem cells. Further study is required to understand how the interaction between 14-3-3σ and KCMF1 proteins affects cell proliferation and colony formation of SW1116csc.
In summary, we constructed a 14-3-3σ bait gene and successfully expressed it as a fusion with the GAL4 DNA-binding domain. Using the yeast two-hybrid system, we found novel binding proteins (KCMF1, NQO2, HIBADH and 14-3-3σ) from the HeLa cDNA library that closely interact with 14-3-3σ. The knock-down of 14-3-3σ and KCMF1 protein expression significantly inhibited cell proliferation and colony formation of colon cancer stem cells.
The cancer stem cell (CSC) hypothesis is currently at the center of a rapidly evolving field, involving a change of perspective on the development and treatment of cancers. However, research has been hampered by the lack of distinct molecular markers of CSCs. Among all 14-3-3 proteins, 14-3-3σ is the isoform most directly linked to cancer. There are several lines of evidence indicating that 14-3-3σ acts as a tumor suppressor gene and that its inactivation is crucial in tumorigenesis.
All 14-3-3 proteins bind to phosphoserine/phosphothreonine-containing peptide motifs corresponding to the sequences RSXpSXP or RXXXpSXP. Many 14-3-3-binding proteins contain sequences that closely match these motifs, although a number of ligands bind to 14-3-3 in a phospho-independent manner using alternative sequences that do not closely resemble these motifs. The interaction of 14-3-3σ with other proteins may be involved in proliferation and colony formation of human colon cancer stem cells.
The authors constructed 14-3-3σ bait gene and expressed as a fusion to the GAL4 DNA-binding domain successfully. Using the yeast two-hybrid system, we found novel binding proteins [potassium channel modulatory factor 1 (KCMF1), quinone oxidoreductase, hydroxyisobutyrate dehydrogenase and 14-3-3σ] from the HeLa cDNA library which closely interact with 14-3-3σ. The knock-down expression of 14-3-3σ and KCMF1 proteins significantly inhibited cell proliferation and colony formation of colon cancer stem cells.
The study of CSCs has important implications for future cancer treatment and therapies. The CSC hypothesis states that if the CSCs were eliminated, the tumor would simply regress due to differentiation and cell death. By selectively targeting CSCs relative proteins, it may be possible to treat patients with aggressive, non-resectable tumors and prevent the tumor from metastasizing.
14-3-3 proteins are among the most abundant proteins within the cell, having been initially identified in 1967 as a family of acidic proteins within the mammalian brain. This family of highly conserved proteins consisting of seven isotypes in human cells (β, γ, ε, η, σ, τ, ξ) plays crucial roles in regulating multiple cellular processes including the maintenance of cell cycle checkpoints and DNA repair, the prevention of apoptosis, the onset of cell differentiation and senescence, and the coordination of cell adhesion and motility.
The paper presents an original work about colon CSCs, and shows an original pathway for colon cancer management.
P- Reviewer Torigoe T S- Editor Zhai HH L- Editor A E- Editor Li JY
| 1. | Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1237] [Cited by in RCA: 1299] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 2. | Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 368] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 3. | Ferl RJ, Manak MS, Reyes MF. The 14-3-3s. Genome Biol. 2002;3:REVIEWS3010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA. 2000;97:6049-6054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 348] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 5. | Lodygin D, Yazdi AS, Sander CA, Herzinger T, Hermeking H. Analysis of 14-3-3sigma expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene. 2003;22:5519-5524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Lodygin D, Diebold J, Hermeking H. Prostate cancer is characterized by epigenetic silencing of 14-3-3sigma expression. Oncogene. 2004;23:9034-9041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Yatabe Y, Osada H, Tatematsu Y, Mitsudomi T, Takahashi T. Decreased expression of 14-3-3sigma in neuroendocrine tumors is independent of origin and malignant potential. Oncogene. 2002;21:8310-8319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Moreira JM, Gromov P, Celis JE. Expression of the tumor suppressor protein 14-3-3 sigma is down-regulated in invasive transitional cell carcinomas of the urinary bladder undergoing epithelial-to-mesenchymal transition. Mol Cell Proteomics. 2004;3:410-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 sigma gene is associated with 5’ CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353-4357. [PubMed] |
| 10. | Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298-5302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 189] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 11. | Zou J, Yu XF, Bao ZJ, Dong J. Proteome of human colon cancer stem cells: a comparative analysis. World J Gastroenterol. 2011;17:1276-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Hammond NL, Headon DJ, Dixon MJ. The cell cycle regulator protein 14-3-3σ is essential for hair follicle integrity and epidermal homeostasis. J Invest Dermatol. 2012;132:1543-1553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Thanasopoulou A, Stravopodis DJ, Dimas KS, Schwaller J, Anastasiadou E. Loss of CCDC6 affects cell cycle through impaired intra-S-phase checkpoint control. PLoS One. 2012;7:e31007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Zhang J, Chen F, Li W, Xiong Q, Yang M, Zheng P, Li C, Pei J, Ge F. 14-3-3ζ interacts with stat3 and regulates its constitutive activation in multiple myeloma cells. PLoS One. 2012;7:e29554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22:688-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 224] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 16. | Geng C, Sang M, Yang R, Gao W, Zhou T, Wang S. Overexpression of 14-3-3σ counteracts tumorigenicity by positively regulating p73 in vivo. Oncol Lett. 2011;2:1177-1182. [PubMed] |
| 17. | Inglés-Esteve J, Morales M, Dalmases A, Garcia-Carbonell R, Jené-Sanz A, López-Bigas N, Iglesias M, Ruiz-Herguido C, Rovira A, Rojo F. Inhibition of specific NF-κB activity contributes to the tumor suppressor function of 14-3-3σ in breast cancer. PLoS One. 2012;7:e38347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Ling C, Su VM, Zuo D, Muller WJ. Loss of the 14-3-3σ tumor suppressor is a critical event in ErbB2-mediated tumor progression. Cancer Discov. 2012;2:68-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 166] [Reference Citation Analysis (0)] |
| 19. | Hynes NE, Smirnova T. The 14-3-3σ tumor suppressor has multiple functions in ErbB2-induced breast cancer. Cancer Discov. 2012;2:19-22. [PubMed] |
| 20. | Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3sigma functional specificity. J Biol Chem. 2005;280:18891-18898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 193] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 21. | Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 22. | Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1050] [Cited by in RCA: 1093] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 23. | Urschel S, Bassermann F, Bai RY, Münch S, Peschel C, Duyster J. Phosphorylation of grb10 regulates its interaction with 14-3-3. J Biol Chem. 2005;280:16987-16993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Benzinger A, Muster N, Koch HB, Yates JR, Hermeking H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics. 2005;4:785-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436-1450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 366] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 26. | Harada S, Fujii C, Hayashi A, Ohkoshi N. An association between idiopathic Parkinson’s disease and polymorphisms of phase II detoxification enzymes: glutathione S-transferase M1 and quinone oxidoreductase 1 and 2. Biochem Biophys Res Commun. 2001;288:887-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Iskander K, Paquet M, Brayton C, Jaiswal AK. Deficiency of NRH: quinone oxidoreductase 2 increases susceptibility to 7,12-dimethylbenz(a)anthracene and benzo(a)pyrene-induced skin carcinogenesis. Cancer Res. 2004;64:5925-5928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Gaikwad NW, Yang L, Rogan EG, Cavalieri EL. Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones. Free Radic Biol Med. 2009;46:253-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Gong X, Kole L, Iskander K, Jaiswal AK. NRH: quinone oxidoreductase 2 and NAD(P)H: quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67:5380-5388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 30. | Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, van Vliet MH, Wessels LF, Peterse JL. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104:12111-12116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 371] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 31. | Aprelikova O, Pace AJ, Fang B, Koller BH, Liu ET. BRCA1 is a selective co-activator of 14-3-3 sigma gene transcription in mouse embryonic stem cells. J Biol Chem. 2001;276:25647-25650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Li Z, Stuart RO, Eraly SA, Gittes G, Beier DR, Nigam SK. Debt91, a putative zinc finger protein differentially expressed during epithelial morphogenesis. Biochem Biophys Res Commun. 2003;306:623-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 33. | Jang JH. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett. 2004;578:21-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Kreppel M, Aryee DN, Schaefer KL, Amann G, Kofler R, Poremba C, Kovar H. Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing’s sarcoma cells. Oncogene. 2006;25:2795-2800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 35. | Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649-2657. [PubMed] |