Published online Apr 14, 2013. doi: 10.3748/wjg.v19.i14.2197
Revised: February 2, 2013
Accepted: February 8, 2013
Published online: April 14, 2013
Processing time: 108 Days and 3.8 Hours
AIM: To investigate the regulative effect of miRNA-338-3p (miR-338-3p) on cell growth in colorectal carcinoma (CRC).
METHODS: The lentiviral vector pLV-THM-miR-338-3p and pLV-THM-miR-338-3p-inhibitor were constructed. The recombinant viral vector encoding the pre-miR-338-3p or miR-338-3p-inhibitor and the two packaging plasmids psPAX2 and pMD2.G were cotransfected into human embryonic kidney 293T cells to package lentivirus. The supernatant containing the lentivirus particles was harvested to determine the viral titer, and this supernatant was then used to transduce CRC-derived cell line, SW-620. Flow cytometry was utilized for sorting the green fluorescent protein (GFP)+ cells to establish the SW-620 cell line stably expressing pre-miR-338-3p or miR-338-3p-inhibitor. Moreover, the expression of miR-338-3p was determined by real-time reverse transcriptase polymerase chain reaction, and Western blotting was used to detect the expression of the smoothened (SMO, the possible target of miR-338-3p) protein in SW-620 cells. Furthermore, the status of CRC cell proliferation and apoptosis were detected by 3-(4,5-dimethyl-2 thiazoyl)-2,5-diphenyl-2H-tetrazolium bromide assay and flow cytometry, respectively.
RESULTS: Restriction enzyme digestion and DNA sequencing demonstrated that the lentiviral vector pLV-THM-miR-338-3p and pLV-THM-miR-338-3p-inhibitor were constructed successfully. GFP was expressed after the SW-620 cells were transduced by the lentivirus. Expression of miR-338-3p in SW-620 cells transduced with the lentivirus pLV-THM-miR-338-3p was significantly increased (relative expression 3.91 ± 0.51 vs 2.36 ± 0.44, P < 0.01). Furthermore, overexpression of miR-338-3p inhibited the expression of SMO protein in SW-620 cells, which showed obviously suppressed proliferation ability [cellular proliferation inhibition rate (CPIR) 61.9% ± 5.2% vs 41.6% ± 4.8%, P < 0.01]. Expression of miR-338-3p in SW-620 cells transduced with the lentivirus pLV-THM-miR-338-3p-inhibitor was significantly decreased (relative expression 0.92 ± 0.29 vs 2.36 ± 0.44, P < 0.01). Moreover, the downregulated expression of miR-338-3p caused upregulated expression of the SMO protein in SW-620 cells, which showed significantly enhanced proliferation ability (CPIR 19.2% ± 3.8% vs 41.6% ± 4.8%, P < 0.01). However, anti-SMO-siRNA largely, but not completely, reversed the effects induced by blockage of miR-338-3p, suggesting that the regulative effect of miR-338-3p on CRC cell growth was indeed mediated by SMO.
CONCLUSION: miR-338-3p could suppress CRC growth by inhibiting SMO protein expression.
Core tip: The previous study has shown that loss of miR-338-3p expression is associated with clinical aggressiveness of colorectal carcinoma (CRC). In this study, the authors demonstrated that forced expression of miR-338-3p in CRC cells suppressed cell proliferation and induced apoptosis, whereas inhibition of miR-338-3p in CRC cells promoted growth. We described miR-338-3p as a direct regulator of smoothened (SMO) expression in CRC, showing a new mechanism responsible for SMO upregulation in CRC. This study provides evidence for antiangiogenic activity of miR-338-3p in the development of CRC and it may develop as a useful biomarker or therapeutic target in CRC.
- Citation: Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. miRNA-338-3p suppresses cell growth of human colorectal carcinoma by targeting smoothened. World J Gastroenterol 2013; 19(14): 2197-2207
- URL: https://www.wjgnet.com/1007-9327/full/v19/i14/2197.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i14.2197
Colorectal carcinoma (CRC) is one of the leading causes of cancer-related death worldwide with an estimated one million new cases and 500 000 deaths annually. The CRC incidence and mortality in China have increased rapidly in the past few decades[1]. Screening for CRC allows early-stage diagnosis of the malignancy and potentially reduces mortality. New targeted therapies directed against molecules involved in the pathogenesis of CRC have recently been reported to be safe and effective[2,3]. With the advent of new chemotherapeutic agents, such as angiogenesis inhibitor and transforming growth factor-α inhibitors, there is growing interest to identify new prognostic biomarkers and therapeutic targets for this disease[4].
miRNAs are a new class of small noncoding RNAs that regulate the expression of target genes through translational repression or mRNA cleavage/decay[5,6]. Genome-wide studies have demonstrated that miRNA genes are frequently located at cancer-associated genomic regions or in fragile sites, and in minimal regions of loss of heterozygosity or of amplification, or in common breakpoint regions, indicating the potential roles of miRNAs in tumorigenesis[7,8]. miRNAs have been demonstrated to play an important role in the multi-step processes of carcinogenesis, either by oncogenic or tumor suppressor function[9]. Studies of miRNAs have been extended to many types of tumors, including CRC. These studies have revealed that miRNAs may be potential diagnostic or prognostic tools for cancer, and the identification of target mRNAs is a key step for assessing the role of aberrantly expressed miRNAs in human cancer[10].
miR-338-3p has recently been discovered and is involved in cell growth. Although miR-338-3p is known to be specifically expressed in neuronal tissue, little is known about its abundance and function during carcinogenesis[11,12]. We have found that miR-338-3p is downregulated in several CRC samples compared with adjacent non-tumorous tissues, suggesting that miR-338-3p might act as tumor suppressor in CRC, however, the targets that it regulates in CRC have not been established. Smoothened (SMO) protein is related to G-protein-coupled receptors, and is the key activator of the Hedgehog (Hh) signaling pathway[13,14]. Upregulation of SMO in CRC is correlated with higher biological aggressiveness, advanced stage, poor differentiation, larger tumor size, and high proliferative activity[15]. Furthermore, it is also well known that SMO regulation, both in physiological and pathological conditions, is mostly at a post-transcriptional level[16]. Moreover, with the application of bioinformatics predictions, we have found that miR-338-3p and SMO mRNA 3’-untranslated region (UTR) have complementary binding sites. Thus, we inferred that the noncoding RNA, miR-338-3p, acts as a local regulator of SMO by binding to the 3’-UTR of its mRNA, thereby modulating CRC development. In order to verify this hypothesis, we investigated the regulative effect of miR-338-3p on cell proliferation and apoptosis in CRC. We aimed to reveal a new regulatory mechanism of miR-338-3p in the development of CRC, and provide a new miRNA and target gene for clinical application.
The lentiviral vectors used in this study were pLV-THM, psPAX2, and pMD2.G, which were a transfer vector, packaging plasmid, and envelope plasmid, respectively. The sequences of interest were inserted into the transfer vector between the MluI and ClaI restriction sites according to the Addgene protocol. The third generation of self-inactivating, lentivirus plasmid, pLV-THM (HIV-1-based vector; Addgene, Cambridge, MA, United States), which contains a CMV-driven enhanced green fluorescence protein (GFP) reporter and an H1 promoter upstream of the restriction sites (MluI and ClaI), was used as the transfer plasmid and was linearized by digesting the vector with the restriction enzymes. The sequence of the mature miR-338-3p (5’-UCCAGCAUCAGUGAUUUUGUUG-3’) was obtained from miRBase (http://www.mirbase.org/). The pre-miR-338-3p and miR-338-3p-inhibitor oligonucleotides were chemically synthesized by Sangon Biotech Co. Ltd. (Shanghai, China) and were inserted between the MluI and ClaI sites of the pLV-THM plasmid. After the pre-miR-338-3p and miR-338-3p-inhibitor lentiviral-based vector were transformed into competent Escherichia coli DH5α cells using the calcium chloride method, antibiotic-resistant colonies were selected on LB-ampicillin agar plates. After colony selection and further propagation, the plasmid was extracted using the alkaline lysis method. The plasmid DNA was then analyzed by restriction enzyme digestion and sequence analysis. The plasmid containing the target gene was digested with the restriction enzymes and amplified by polymerase chain reaction (PCR). The clones with positive PCR results were subjected to DNA sequencing.
Human embryonic kidney 293T (HEK-293T) cells (Invitrogen, Carlsbad, CA, United States) and the human CRC-derived cell line SW-620 (Shanghai Institutes for Biological Science, CAS, China) were cultured in Dulbecco’s Modified Eagle’s Medium high glucose supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT, United States) at 37 °C in a humidified incubator with 5% CO2. The medium was changed every 3 d, and the cells were trypsinized with trypsin/ethylene diamine tetraacetic acid when 80%-90% confluence was reached. Cells at passages 4-8 were used for the experiments.
Twenty-four hours prior to transfection, the HEK-293T cells in logarithmic growth phase were trypsinized, and the cell density was adjusted to 1.0 × 106 cells/mL with complete culture medium. The cells were reseeded into 15-cm cell culture dishes and cultured for 24 h prior to transfection. The cells were 90%-95% confluent on the day of transfection. The recombinant viral vector encoding the miR-338-3p or miR-338-3p-inhibitor and the two packaging plasmids psPAX2 and pMD2.G were extracted with a plasmid extraction kit (Invitrogen) and cotransfected into HEK-293T cells according to the manufacturer’s instructions. After 8 h transfection, the cell culture medium was replaced with fresh complete medium. After 24 h transfection, the expression of GFP was determined. After 48 h transfection, the culture medium was collected and centrifuged at 4000 ×g at 4 °C for 10 min to remove any cellular debris. The supernatant was filtered through a 0.45-μm filter into a Plus-20 centrifugal ultrafiltration unit and centrifuged at 4000 ×g to obtain a high-titer lentivirus stock. The lentivirus without the transgene was used as the negative control and was produced in the same manner.
SW-620 cells were seeded at 1.0 × 105 cells per well in 24-well plates in DMEM containing 10% FBS. After 24 h incubation, the cells were transduced with each lentivirus stock (3.0 × 105 Titer Units). The SW-620 cells were then incubated for an additional 48-72 h prior to identifying the GFP+ cells by flow cytometry (Becton Dickinson, San Jose, CA, United States).
Total RNA from SW-620 cells was prepared using the TRIzol reagent (Invitrogen) after viral transduction. The precipitate was dissolved in diethylpyrocarbonate-treated water, and a nucleic acid protein analyzer (Beckman Coulter, Fullerton, CA, United States) was used to determine the RNA concentration. The purity and integrity of the RNA were identified as follows: the A260nm/A280nm was ≥ 1.8, and the band ratio of 28 S RNA to 18 S RNA was ≥ 1.5 in formaldehyde denaturing gel electrophoresis. Accurate quantitation of the mature miR-338-3p was obtained using the TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, United States). The reverse transcription reaction was performed using 10 ng total RNA and the looped primers. Real-time PCR was performed using the standard TaqMan MicroRNA Assays protocol on the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). The PCR reaction (20 μL) included 1.33 μL reverse transcription product, 1 × TaqMan Universal PCR Master Mix, No AmpErase UNG, 0.2 μmol/L TaqMan probe, 1.5 μmol/L forward primer, and 0.7 μmol/L reverse primer. The reactions were incubated in a 96-well plate at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The miR-338-3p expression level was measured using the Ct (threshold cycle) method. Ct is the fractional cycle number at which the fluorescence of each sample passes the fixed threshold. The ΔΔCT method for relative quantitation of gene expression was used to determine the miR-338-3p expression levels. The ΔCT was calculated by subtracting the Ct of U6 from the Ct of the miR-338-3p. The ΔΔCT was calculated by subtracting the ΔCT of the reference sample from the ΔCT of each sample. The fold change was calculated using the equation 2-ΔΔCT. The TaqMan MicroRNA Assays for U6 RNA was used to normalize the relative abundance of miR-338-3p.
The analysis of miR-338-3p-predicted targets was performed using the algorithms TargetScan (http://targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and MiRanda (http://www.microrna.org/microrna/home.do).
SW-620 cells were rinsed twice with cold PBS and were then lysed in ice-cold lysis buffer containing 150 mmol/L NaCl, 50 mmol/L Tris-HCl (pH 7.6), 0.1% SDS, 1% Nonidet P-40, and protease inhibitor cocktail (Boehringer Mannheim, Lewes, United Kingdom). The samples were cleared by centrifugation at 13 000 ×g for 10 min. The cellular protein (50 μg) was subjected to SDS-PAGE and electrotransferred to polyvinylidine fluoride membranes (Immobilon, Bedford, MA, United States). After blocking in 20 mmol/L Tris-HCl, (pH 7.6) containing 150 mmol/L NaCl, 0.1% Tween-20, and 5% nonfat dry milk, the membranes were incubated with primary antibodies against SMO or β-actin, which was used as a sample loading control, overnight at 4 °C. The membranes were then incubated with horseradish-peroxidase-conjugated secondary antibody. The blot was developed using the ECL detection kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ, United States) according to the manufacturer’s instructions.
The status of cell proliferation was determined by 3-(4,5-dimethyl-2 thiazoyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Amresco, Solon, OH, United States) assay. Exponentially growing SW-620 cells were adjusted to 2.5 × 104 cells/mL with DMEM, plated in 96-well plates (Corning, Corning, NY, United States) at 200 μL/well and then incubated for 12 h according to routine procedure. After being transduced with each lentivirus stock and incubated for 48 h (5 duplicate wells for each sample), 20 μL/well MTT (5 g/L) was added to each well. The medium was then removed after 4 h incubation and 100 μL/well dimethyl sulfoxide was added to dissolve the reduced formazan product. Finally, the plate was read in an enzyme-linked immunosorbent microplate reader (Bio-Rad 2550) at 490 nm. The cellular proliferation inhibition rate (CPIR) was calculated using the following formula: CPIR = (1 - average A value of experimental group/average A value of control group) × 100%.
The effects of miR-338-3p on CRC cell cycle and apoptosis were examined by flow cytometry. Pretreated SW-620 cells were harvested and washed twice with PBS, fixed with 70% ethanol at -20 °C for 30 min, and stored at 4 °C overnight, then washed with PBS again, treated with 100 mL 100 mg/L RNase at 37 °C for 30 min, and stained with 100 mL 50 mg/L propidium iodide at 4 °C for 30 min in the dark. The multiplication cycle and apoptotic rate were assayed using flow cytometry, and the data were analyzed using CellQuest software. The percentages of cells in the G0/G1 phase and S phase, and the apoptotic rate were measured by calculating the ratio of the number of corresponding cells to the number of total cells. For each sample, 10 000 cells were measured.
The relative expression analysis of the target gene was performed using REST-XL (Relative Expression Software Tool, available at http://www.wzw.tum.de/genequantification). All data in the experiment were presented as the mean ± SD. Comparisons between the groups were analyzed with one-way ANOVA and Student-Newman-Keuls Q test, using SPSS version 13.0 software (SPSS Inc., Chicago, IL, United States). P < 0.05 was considered statistically significant.
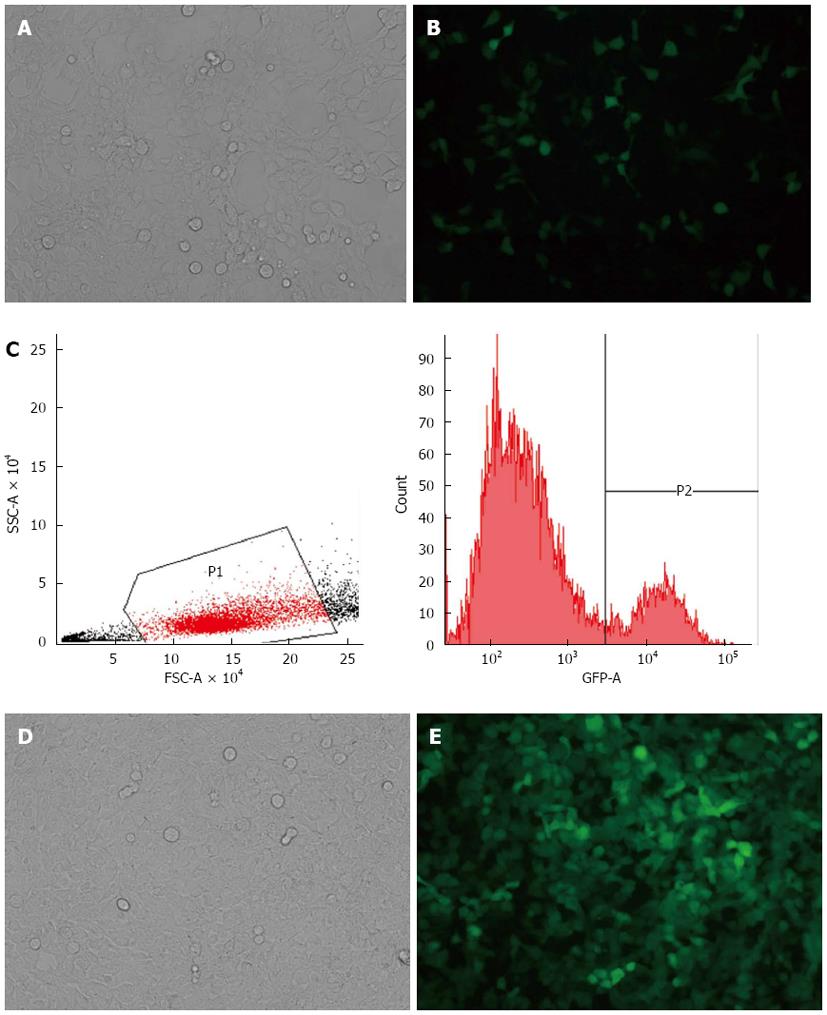
HEK-293T cells were cotransfected with the transfer plasmid, pLV-THM-transgene, the packaging plasmid, psPAX2, and the envelope plasmid, pMD2.G. The high-titer lentivirus was harvested as the stock virus solution. GFP was expressed 48 h after the SW-620 cells were transduced by the lentivirus, and the cells were observed under a fluorescence microscope (Figure 1A, B). This suggests that the miR-338-3p or miR-338-3p-inhibitor vector was successfully transduced into the SW-620 cells, which provides the basis for further studies regarding the molecular function of miR-338-3p in CRC cells. The GFP+ fluorescent cells were then identified and harvested using flow cytometry for the next experiment (Figure 1C-E).
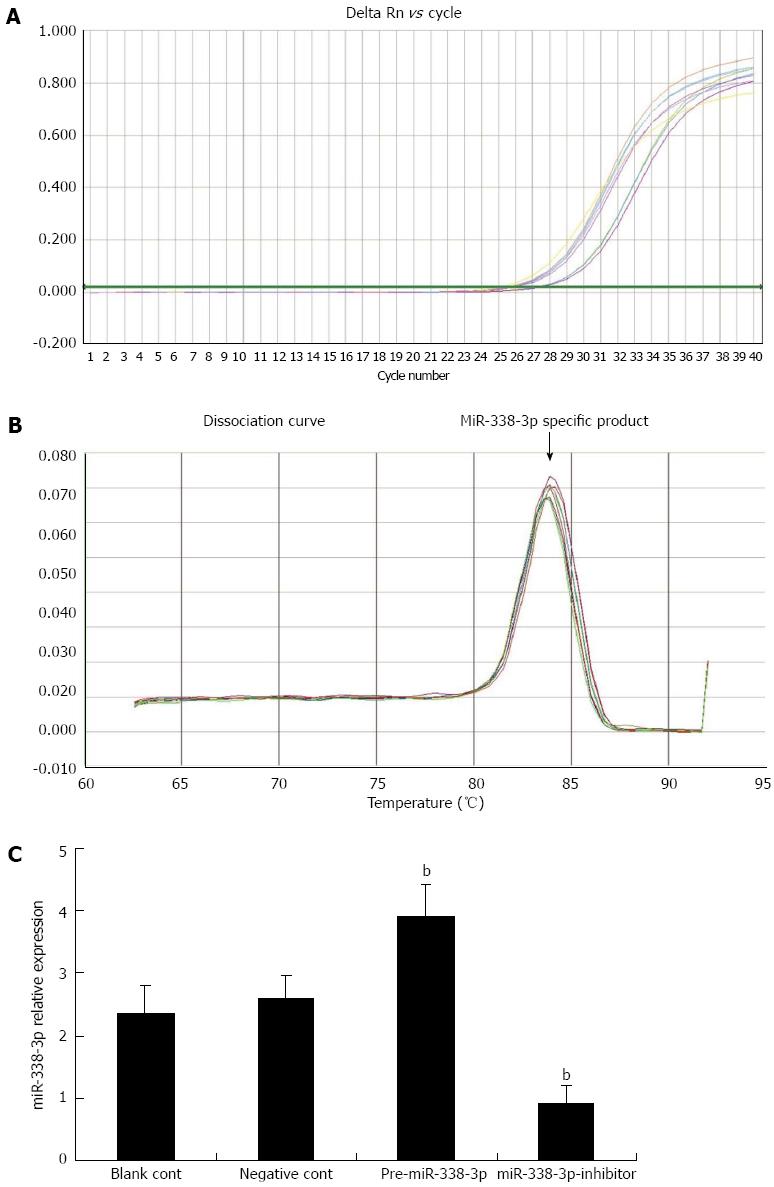
To study the expression pattern of miR-338-3p in SW-620 cells after lentivirus transduction, we performed real-time reverse transcriptase (RT)-PCR to detect miR-338-3p expression in the SW-620 cells. Real-time RT-PCR indicated that the miR-338-3p cDNA increased exponentially and then reached a plateau. The miR-338-3p amplification curve was a typical reverse S pattern (Figure 2A) and showed higher amplification efficiency. The miR-338-3p PCR product was 72 bp long, the corresponding Tm was 84.09 ± 0.15 °C, the melting temperature was even, and the shape of the peak was sharp (Figure 2B). As shown in Figure 2C, the expression level of miR-338-3p in the pLV-THM-miR-338-3p group was more than one-third of the expression in the control cells that were transduced with the blank pLV-THM vector, whereas the expression level of miR-338-3p in the pLV-THM-miR-338-3p-inhibitor group decreased significantly compared with the control group (P < 0.01). Thus, we established the SW-620-miR-338-3p and SW-620-miR-338-3p-inhibitor cell lines successfully to observe the corresponding biological effect.
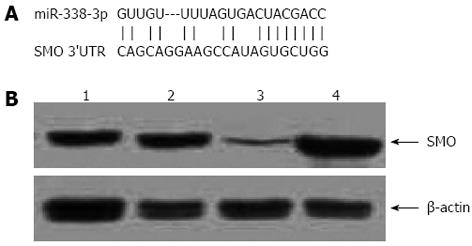
Most miRNAs are thought to control gene expression by base-pairing with the miR-recognizing elements found in their messenger target. We then used all three currently available major prediction programs, including TargetScan, Miranda and PicTar, to analyze the potential interaction between miR-338-3p and SMO. SMO mRNA was predicted by all of the algorithms and revealed potential miR-338-3p target sites in its 3’-UTR (Figure 3A).
To check if miR-338-3p actually affected SMO expression in CRC cells, we analyzed the consequence of the ectopic expression of miR-338-3p. We transfected the pre-miR-338-3p and miR-338-3p-inhibitor into SW-620 cells by lentivirus transduction as described above, and we searched for changes in SMO protein levels by Western blotting analysis. Introduction of pre-miR-338-3p caused a significant increase of miR-338-3p value and decreased SMO protein levels in SW-620 cells. Conversely, miR-338-3p-inhibitor caused a significant decrease of miR-338-3p value and increased SMO protein level (Figure 3B). This result strongly validates a post-transcriptional regulation of SMO protein by miR-338-3p.
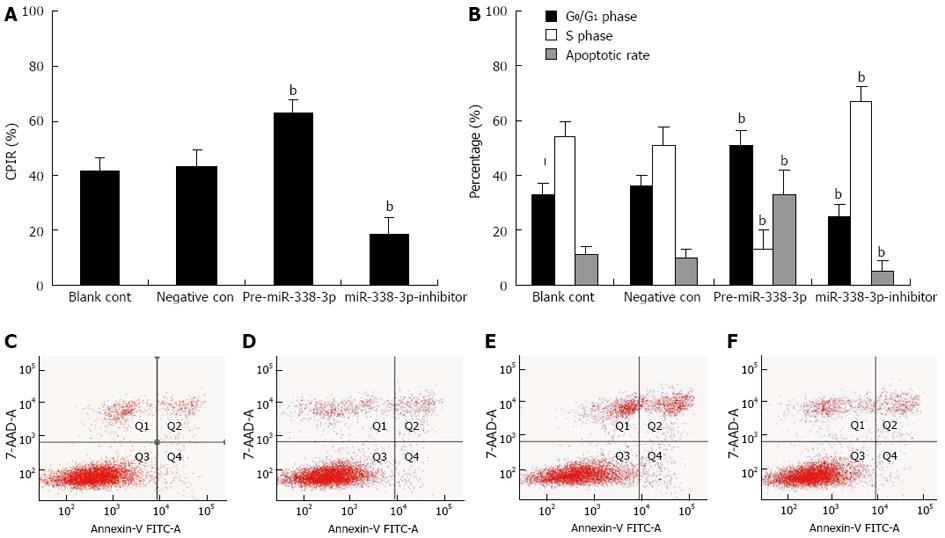
SMO has a key role in the cell cycle, particularly in the growth arrest at the G1/S transition, therefore, we further tested if the cell growth potential of stably transduced CRC cells expressing miR-338-3p or miR-338-3p-inhibitor was modified as a consequence of the demonstrated SMO alteration. First, to evaluate the effect of miR-338-3p on CRC cell proliferation, growing SW-620 cells were transduced with lentivirus pLV-THM-miR-338-3p or pLV-THM-miR-338-3p-inhibitor for 48 h and the cell proliferation was determined by MTT assay. We observed a significant increase in proliferation after transduction of pLV-THM-miR-338-3p-inhibitor (Figure 4A, P < 0.01). In contrast, pre-miR-338-3p significantly inhibited cell proliferation (Figure 4A, P < 0.01). These data indicate that cell proliferation can be significantly suppressed by increased miR-338-3p expression. Second, we performed flow cytometry analysis after exposure to miR-338-3p or miR-338-3p-inhibitor to investigate CRC cell-cycle phase distribution. SW-620 cells overexpressing miR-338-3p had a significant decrease in the S-phase population and a increase in the G0/G1 population compared with cells transduced with negative control lentivirus (Figure 4B, P < 0.01). On the contrary, miR-338-3p-inhibitor significantly increased the S-phase and decreased the G0/G1 population (Figure 4B, P < 0.01). Third, we investigated the effect of miR-338-3p on apoptosis by flow cytometry and found that apoptosis increased dramatically in SW-620 cells after transduction with lentivirus pLV-THM-miR-338-3p, suggesting that miR-338-3p may function as a strong apoptotic inducer in human CRC cells (Figure 4C-F). These results confirm the potential tumor-suppressor activity of miR-338-3p in CRC.
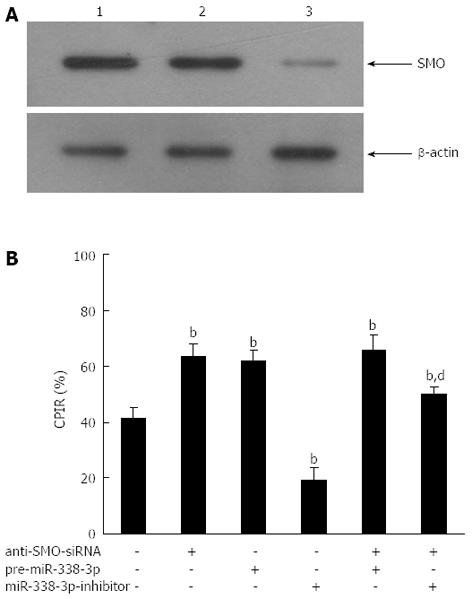
If miR-338-3p suppression of CRC cell proliferation was indeed mediated by SMO, we would expect that the SMO-specific and irreversible antagonist anti-SMO-siRNA would abolish this effect. To test this hypothesis, we measured the changes in proliferation induced by pre-miR-338-3p or miR-338-3p-inhibitor in CRC cells previously transfected with anti-SMO-siRNA. The aim was to study if and how the SMO-depleted cellular environment responded to pre-miR-338-3p or miR-338-3p-inhibitor. SW-620 cells were pretreated with or without anti-SMO-siRNA (50 nmol/L) for 24 h prior to transduction with lentivirus pLV-THM-miR-338-3p or pLV-THM-miR-338-3p-inhibitor, and cell proliferation was determined by MTT assay. A reduction in SMO level, by means different from miR-338-3p overexpression, led to analogous outcomes. When we transfected SW-620 cells with anti-SMO-siRNA, SMO protein was reduced by about 80% (Figure 5A), and we observed a sharp decrease in cell proliferation as compared with negative controls (Figure 5B, P < 0.01). Thus, reducing SMO protein levels in CRC cells, either by miR-338-3p overexpression or by anti-SMO-siRNA transfection, is sufficient to induce a comparable decrease in cell growth.
When lentivirus pLV-THM-miR-338-3p was transduced into SW-620 cells previously treated with anti-SMO-siRNA, we observed that anti-SMO-siRNA and pre-miR-338-3p seemed to co-operate to inhibit the growth rate (Figure 5B). However, when lentivirus pLV-THM-miR-338-3p-inhibitor was transduced into SW-620 cells previously treated with anti-SMO-siRNA, we observed that the enhancement of cell proliferation by miR-338-3p-inhibitor was largely abrogated by anti-SMO-siRNA (Figure 5B, P < 0.01). These results indicated that the promotive effect of miR-338-3p-inhibitor on CRC cell growth was largely, but not completely, mediated by SMO, suggesting that miR-338-3p-inhibitor could also activate some SMO-independent signaling pathway to promote CRC cell growth in addition to upregulation of SMO.
With the advent of new chemotherapeutic agents, clarification of the molecular pathogenesis of CRC is crucial for developing effective therapeutic strategies to improve patient outcome[17,18]. The miR-338 gene is located on chromosome 17 and produces two mature forms (miR-338-3p and miR-338-5p). Tsuchiya et al[19] have reported that miR-338-3p contributes to the formation of epithelial basolateral polarity by facilitating the translocalization of β1-integrin to the basolateral membrane, which highlights a potentially important role for miR-338-3p in the process of epithelial cell differentiation. Huang et al[20] have demonstrated that a decrease in miR-338-3p expression in hepatocellular carcinoma, which is another type of epithelial-cell-derived cancer, was significantly associated with TNM stage, vascular invasion, intrahepatic metastasis, tumor size, and tumor grade. Our previous study has also shown that loss of miR-338-3p expression is associated with clinical aggressiveness of CRC. Moreover, the miR-338-3p expression was not only related to TNM stage but also to tumor invasion and migration. The level of miR-338-3p expression at TNM stages III and IV was lower than that at stages I and II, and the tumors which invaded adjacent tissues or organs had less miR-338-3p expression than those limited to the wall of the colon and rectum (data not shown). Thus, miR-338-3p may be an important tumor suppressor, which can cleave or inhibit the targeted mRNAs of tumor promoters, and play a role in the progression of CRC. However, lack of knowledge about the targets for miR-338-3p hampers a full understanding of the biological functions deregulated by miR-338-3p aberrant expression. To confirm the molecular mechanism of miR-338-3p in CRC, it is necessary to observe the biological effects of the up- and down-regulation of miR-338-3p. Thus, we constructed a CRC-derived cell line in which miR-338-3p was stably over- or under-expressed by transducing the lentivirus vector, pLV-THM-miR-338-3p or pLV-THM-miR-338-3p-inhibitor, into SW-620 cells[21]. Notably, we showed that our combined lentivirus specifically enhanced or inhibited endogenous miR-338-3p to induce the corresponding biological effect. The successful construction of the lentiviral vector provides the basis for further studies regarding the molecular function of miR-338-3p in CRC[22-24].
To extend our previous observation, we focused on the role of miR-338-3p in regulation of proliferation and apoptosis in CRC. We found that the proliferative potential was suppressed after restoration of miR-338-3p expression in CRC cells transduced by lentivirus vector, pLV-THM-miR-338-3p. However, the downregulation of miR-338-3p, due to transducing by lentivirus vector pLV-THM-miR-338-3p-inhibitor into SW-620 cells, induced CRC cell proliferation. Cell cycle status and apoptosis are usually closely associated. Cells failing to progress to mitosis are destined for apoptosis. Besides cell-cycle arrest, the inhibition of cell growth observed in CRC cells with pre-miR-338-3p may also be a result of increased apoptosis. In this study, treatment of lentivirus pLV-THM-miR-338-3p caused G0/G1 phase arrest and blocked cells from entering S phase. Interestingly, as seen in other tumor cells, we clearly demonstrated that pre-miR-338-3p induced significant apoptosis in CRC cells, as demonstrated by flow cytometry. These data demonstrate that miR-338-3p is a potential tumor suppressor for CRC. However, the exact mechanisms of miR-338-3p remain unknown.
With the application of bioinformatics prediction programs, such as TargetScan, PicTar and MiRanda, we found that miR-338-3p and the 3’-UTR of SMO mRNA had complementary binding sites. From this, we hypothesized that SMO may be a new target of miR-338-3p in CRC; however, this finding has not yet been reported. SMO, a seven-membrane-spanning receptor is a fundamental component of the Hh signaling pathway and an important anticancer drug target[25-27]. Once activated, SMO triggers a series of intracellular events with resultant activation of the zinc finger transcription effectors including Gli, which in turn regulates cell proliferation, differentiation, apoptosis and invasion[28-30]. It has been reported that 3-Keto-N-(aminoethyl-aminocaproyl-dihydrocinnamoyl) cyclopamine (KAAD-cyclopamine), a synthetic specific antagonist of SMO, markedly inhibits hepatocellular carcinoma cell growth and motility by binding to SMO[31]. Indeed, in our study, downregulation of SMO occurred in response to lentivirus vector pLV-THM-miR-338-3p transduction into CRC cells, and significant upregulation of SMO occurred in response to lentivirus vector pLV-THM-miR-338-3p-inhibitor transduction. Consistent with Huang et al[32], our results suggest that SMO is a direct target of miR-338-3p in CRC cells.
We deduced that miR-338-3p inhibited CRC cell proliferation, likely through downregulating SMO. To confirm this, we performed RNA interference to knock down SMO in CRC cells before transduction with miR-338-3p-inhibitor. We showed that anti-SMO-siRNA could significantly, but not completely, inhibit miR-338-3p-inhibitor-induced proliferation of CRC cells. These results confirmed that the inhibitory effect of miR-338-3p on CRC cell proliferation was largely, but not completely, mediated by SMO, suggesting that miR-338-3p could regulate other SMO-independent signaling pathways to promote CRC growth. We think that our results, which identify SMO as a target for miR-338-3p in the context of CRC cell line, fit well within a dynamic view of the miRNA-mediated regulation of gene expression. It is well known and widely predicted that the relationship between miRNAs and target mRNAs is not a “one to one” connection, because the same mRNA can be regulated by more than one miRNA, and that the choice of how many and which miRNAs target one 3’-UTR is strongly determined by the specific cellular environment[33-35]. An miRNA that regulates targets playing opposite roles in the control of cell proliferation may act as a tumor suppressor in some cancers and as an oncogene in others, depending on which targets are driving tumorigenesis in that specific cellular milieu[36].
In summary, we have described miR-338-3p as a direct regulator of SMO expression in CRC, showing a new mechanism responsible for SMO upregulation in CRC. These findings further outline the importance of miR-338-3p in CRC carcinogenesis. However, it should be emphasized that our results were generated from cultured CRC cells and that they might not necessarily and comprehensively reflect the situation in vivo[37]. Further experiments, beyond the scope of this study, are required to elucidate the antitumor mechanisms of miR-338-3p in athymic mice.
miRNAs regulate gene expression by mainly binding to the 3’-untranslated region (UTR) of the target mRNAs, leading to mRNA degradation or translation inhibition. miRNAs are aberrantly expressed in various cancers, suggesting that they play a vital role as a novel class of oncogenes or tumor suppressor genes, depending on the targets they regulate.
Colorectal carcinoma (CRC) is one of the most serious malignancies in China. Our previous study has shown that loss of miRNA-338-3p (miR-338-3p) expression is associated with clinical aggressiveness of CRC. In this study, the authors report the regulatory effect of miR-338-3p on proliferation and apoptosis of CRC cells.
Some human miRNAs are consistently deregulated in human cancer, suggesting a role for these genes in tumorigenesis. Authors previous study has also shown that loss of miR-338-3p expression is associated with clinical aggressiveness of CRC. The authors demonstrated that forced expression of miR-338-3p in CRC cells suppressed cell growth, whereas inhibition of miR-338-3p promoted cell growth. Furthermore, smoothened (SMO) was identified as a direct target of miR-338-3p. The antiangiogenic role of miR-338-3p was determined as tumor suppressor.
This study indicates that miR-338-3p suppresses cell growth by targeting the SMO gene in CRC in vitro and miR-338-3p might be a novel potential strategy for CRC treatment.
Most miRNAs are thought to control gene expression by base-pairing with the miR-recognizing elements, 3’-UTR, found in their messenger target. Not surprisingly, with the application of bioinformatics predictions, we find that miR-338-3p and SMO mRNA 3’-UTR has complementary binding sites.
miR-338-3p could suppress CRC growth ability by inhibiting SMO protein expression. This study provides evidence for antiangiogenic activity of miR-338-3p in the development of CRC, and may be developed as a useful biomarker or therapeutic target in CRC.
P- Reviewers Bujanda L, Sagaert X S- Editor Wen LL L- Editor A E- Editor Zhang DN
| 1. | Sun K, Wang W, Zeng JJ, Wu CT, Lei ST, Li GX. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;32:375-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 2. | Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18:244-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 223] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 3. | Fabbri M. miRNAs as molecular biomarkers of cancer. Expert Rev Mol Diagn. 2010;10:435-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Li XQ, Guo YY, De W. DNA methylation and microRNAs in cancer. World J Gastroenterol. 2012;18:882-888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Sipos F, Galamb O. Epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions in the colon. World J Gastroenterol. 2012;18:601-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 6. | Reichel M, Li J, Millar AA. Silencing the silencer: strategies to inhibit microRNA activity. Biotechnol Lett. 2011;33:1285-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Lee HC, Kim JG, Chae YS, Sohn SK, Kang BW, Moon JH, Jeon SW, Lee MH, Lim KH, Park JY. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. 2010;136:1073-1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P, Knuutila S. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 9. | Lin M, Chen W, Huang J, Gao H, Ye Y, Song Z, Shen X. MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol Rep. 2011;25:739-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581-12590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 213] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 12. | Luo Y, Zhang S. Computational prediction of amphioxus microRNA genes and their targets. Gene. 2009;428:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Wang K, Pan L, Che X, Cui D, Li C. Sonic Hedgehog/GLI1 signaling pathway inhibition restricts cell migration and invasion in human gliomas. Neurol Res. 2010;32:975-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Coon V, Laukert T, Pedone CA, Laterra J, Kim KJ, Fults DW. Molecular therapy targeting Sonic hedgehog and hepatocyte growth factor signaling in a mouse model of medulloblastoma. Mol Cancer Ther. 2010;9:2627-2636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Stanton BZ, Peng LF. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol Biosyst. 2010;6:44-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | Wang TP, Hsu SH, Feng HC, Huang RF. Folate deprivation enhances invasiveness of human colon cancer cells mediated by activation of sonic hedgehog signaling through promoter hypomethylation and cross action with transcription nuclear factor-kappa B pathway. Carcinogenesis. 2012;33:1158-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 17. | Fang Y, Xiang J, Chen Z, Gu X, Li Z, Tang F, Zhou Z. miRNA expression profile of colon cancer stem cells compared to non-stem cells using the SW1116 cell line. Oncol Rep. 2012;28:2115-2124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, Levy M, Polakova V, Lipska L, Vodicka P. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, Sato F, Tsujimoto G, Shimizu K. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37:3821-3827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Sun K, Guo C, Deng HJ, Dong JQ, Lei ST, Li GX. Construction of lentivirus-based inhibitor of hsa-microRNA-338-3p with specific secondary structure. Acta Pharmacol Sin. 2013;34:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Pan J, Li S, Chi P, Xu Z, Lu X, Huang Y. Lentivirus-mediated RNA interference targeting WWTR1 in human colorectal cancer cells inhibits cell proliferation in vitro and tumor growth in vivo. Oncol Rep. 2012;28:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Li Y, Zhang CY. Analysis of microRNA-induced silencing complex-involved microRNA-target recognition by single-molecule fluorescence resonance energy transfer. Anal Chem. 2012;84:5097-5102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 24. | Haraguchi T, Nakano H, Tagawa T, Ohki T, Ueno Y, Yoshida T, Iba H. A potent 2’-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res. 2012;40:e58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Xu M, Li X, Liu T, Leng A, Zhang G. Prognostic value of hedgehog signaling pathway in patients with colon cancer. Med Oncol. 2012;29:1010-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Arimura S, Matsunaga A, Kitamura T, Aoki K, Aoki M, Taketo MM. Reduced level of smoothened suppresses intestinal tumorigenesis by down-regulation of Wnt signaling. Gastroenterology. 2009;137:629-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | You S, Zhou J, Chen S, Zhou P, Lv J, Han X, Sun Y. PTCH1, a receptor of Hedgehog signaling pathway, is correlated with metastatic potential of colorectal cancer. Ups J Med Sci. 2010;115:169-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Yoshikawa K, Shimada M, Miyamoto H, Higashijima J, Miyatani T, Nishioka M, Kurita N, Iwata T, Uehara H. Sonic hedgehog relates to colorectal carcinogenesis. J Gastroenterol. 2009;44:1113-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Mazumdar T, DeVecchio J, Shi T, Jones J, Agyeman A, Houghton JA. Hedgehog signaling drives cellular survival in human colon carcinoma cells. Cancer Res. 2011;71:1092-1102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 30. | Mazumdar T, Devecchio J, Agyeman A, Shi T, Houghton JA. Blocking Hedgehog survival signaling at the level of the GLI genes induces DNA damage and extensive cell death in human colon carcinoma cells. Cancer Res. 2011;71:5904-5914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Fu X, Yang X, Li J, Tian X, Cai J, Zhang Y. Opposite expression patterns of Sonic hedgehog and Indian hedgehog are associated with aberrant methylation status of their promoters in colorectal cancers. Pathology. 2010;42:553-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Huang XH, Chen JS, Wang Q, Chen XL, Wen L, Chen LZ, Bi J, Zhang LJ, Su Q, Zeng WT. miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J Pathol. 2011;225:463-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 33. | Harquail J, Benzina S, Robichaud GA. MicroRNAs and breast cancer malignancy: an overview of miRNA-regulated cancer processes leading to metastasis. Cancer Biomark. 2012;11:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Pichler M, Winter E, Stotz M, Eberhard K, Samonigg H, Lax S, Hoefler G. Down-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancer. Br J Cancer. 2012;106:1826-1832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 35. | Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Sugihara K, Doki Y. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res. 2012;18:3054-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 36. | Reid JF, Sokolova V, Zoni E, Lampis A, Pizzamiglio S, Bertan C, Zanutto S, Perrone F, Camerini T, Gallino G. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol Cancer Res. 2012;10:504-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 37. | Lévy C, Frecha C, Costa C, Rachinel N, Salles G, Cosset FL, Verhoeyen E. Lentiviral vectors and transduction of human cancer B cells. Blood. 2010;116:498-500; author reply 500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |