Published online Feb 28, 2012. doi: 10.3748/wjg.v18.i8.855
Revised: August 8, 2011
Accepted: August 31, 2011
Published online: February 28, 2012
AIM: To investigate the copy number variation of NACO3 gene in colorectal cancer (CRC) and its correlation with tumor progression.
METHODS: A total of 142 samples of case-matched CRC tissues and adjacent normal tissues were obtained from patients undergoing bowel resection. Quantitative real-time polymerase chain reaction method was used to investigate the copy number variations of NCOA3 as well as gene expression in the collected tissues.
RESULTS: Copy number gains of NCOA3 were detected in 39 CRC samples (27.5%) and were correlated with tumor progression (χ2 = 6.42, P = 0.0112). Moreover, there was a positive correlation between copy number gain and mRNA over-expression of NCOA3 in CRCs (P = 0.0023). Expression level of NCOA3 mRNA was also enhanced in the CRC samples with unaltered copy numbers (3.85 ± 1.23 vs 2.71 ± 0.64, P < 0.01).
CONCLUSION: Sporadic colorectal cancers exhibit different mechanisms of NCOA3 regulation.
-
Citation: Li Z, Fang ZY, Ding Y, Yao WT, Yang Y, Zhu ZQ, Wang W, Zhang QX. Amplifications of
NCOA3 gene in colorectal cancers in a Chinese population. World J Gastroenterol 2012; 18(8): 855-860 - URL: https://www.wjgnet.com/1007-9327/full/v18/i8/855.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i8.855
The p160 steroid receptor coactivator (SRC) family is critical to the transcriptional activation function of nuclear hormone receptors. A key member of this family is nuclear receptor coactivator 3 (NCOA3 and also known as AIB1 and SRC-3), initially found to be amplified and expressed in breast cancer, and has subsequently been shown to express in malignant diseases arising from a wide range of other organs[1-3]. Over-expression of NCOA3 has been implicated in tumor proliferation, invasiveness[4], and resistance to chemoradiotherapy[5] in various kinds of tumors. Increased expression of NCOA3 is also correlated with advanced colorectal carcinoma[6].
Colorectal cancer (CRC) is currently the third most common cancer in Chinese population, responsible for about 130 000 deaths per year. Both genetic and environmental factors contribute to disease etiology, with about one-third of disease variance attributed to inherited genetic factors[7]. The implication of copy-number variations (CNVs) in cancers has become a hot spot over the past few years[8-10]. Studies using SNP arrays and aCGH have suggested that DNA amplification at chromosome position 20q12, also the chromosome locus of NCOA3, is frequent in CRC[11-13]. Moreover, there might be a correlation between CNVs and NCOA3 over-expression in CRCs. However, most of the aCGH experiments focused on the genome-wide screening of CNVs and the data obtained are generally informative but not definitive. Therefore, a study comprehensively examining CNVs in relation to NCOA3 expression or prognosis should be performed using a larger number of tumors.
In the present study, we collected 142 colorectal cancer samples with matched adjacent normal tissues for CNV analysis. Copy number gains of NCOA3 gene were observed in a relatively high percentage of CRC samples and were correlated with cancer lymph node involvement or metastasis. There was also a positive correlation between gene copy number gains and mRNA over-expression of NCOA3. These findings suggested the potential role of CNVs of NCOA3 in CRCs.
CRC samples were obtained from 142 patients undergoing bowel resection at the Department of General Surgery, Henan Province Tumor Hospital. Adjacent normal tissue (ANT) samples located at least 2 cm from the macroscopically unaffected margins of the tumor (polyp or carcinoma) were defined as normal controls. The collected samples were stored in liquid nitrogen. There were 133 adenocarcinomas and 9 mucinous carcinomas (when >50% of the tumor volume was composed of mucin). CRCs were staged according to the Dukes classification system: Dukes A (T1-T2, N0, and M0; n = 42), Dukes B (T3-T4, N0, and M0; n = 37), Dukes C (any T, N1-2, M0; n = 48) and Dukes D (any T and any N and M1; n = 15). Case-matched samples of colorectal carcinomas (n = 142) and normal colonic mucosa (n = 142) were immediately stored in liquid nitrogen after operation. For each sample, a portion of the tissue was disposed to make frozen sections and then micro-dissected by the Eppendorf Microdissection System (Siskiyou MX160L micromanipulator and PixeLink PL-A662 color Charge Coupled Device firewire camera). Then the dissected tissues were subjected to real-time polymerase chain reaction (PCR) analysis. All patients were informed about the aims of specimen collection and gave signed written consent in accordance with the ethical guidelines of Zhengzhou University. Peripheral blood samples from 152 healthy controls were collected at the Department of Histology and Embryology, Basic Medical College of Zhengzhou University. The study was approved by the Ethics Committee of Zhengzhou University.
Genomic DNA was isolated from the tissues using the Genomic DNA Extraction Kit (Innocent, Shenzhen, China) according to the manufacturer’s instructions. The concentration of purified DNA was determined by measuring the absorbance at 260 nm and 280 nm in a spectrophotometer. Pure preparations of DNA have A260/A280 > 1.8. Quantitative PCR was performed using the BioRad Chromo4 real-time PCR system. The primers for RNAse P are: forward: 5’-AGA CTA GGG TCA GAA GCA A-3’ and reverse: 5’-CAT TTC ACT GAA TCC GTT C-3’[14]. The primers for NCOA3 are: forward: 5’-AAA GTA AAC AGA ATG GAT TG-3’ and reverse: 5’-AGT GTG CCT TGG AGT TGA AA-3’. The primer sets for NCOA3 gene were designed by advanced real time PCR primer design tool (GenScript United States Inc.). A melting curve analysis with a temperature gradient of 0.2 K × S–1 from 72 °C to 95 °C was performed following each PCR amplification to confirm that only specific product was amplified.
Average copy numbers of RNAse P in normal candidates (copy numbers = 2) were used as controls[14,15]. The copy numbers of NCOA3 were calculated by the comparative C (T) method[16]. Cut-off values of 0.25, 0.75, 1.25 and 1.75 were used to define the copy numbers as 0, 1, 2 and 3, respectively. Fold change of each sample was presented as follows: fold change = relative expression level/average expression level in the group with 2 copies of DNA.
A standard curve was prepared using 2 μL of crude DNA solutions, including serially diluted samples (original, 2-, 4-, 8-, 16-diluted). The slopes of Ct and efficiency of each primer were calculated by the BioRad Chromo4 real-time PCR system and Microsoft Excel 2007 for Windows. Relative quantification of NCOA3 was performed by the 2-ddCt method[17].
Total RNA was isolated from tissues using the AxyPrepTM Blood Total RNA MiniPrep Kit (Axygen) according to the manufacturer’s instructions. First strand cDNA was synthesized with the RevertAidTM First Stand cDNA Synthesis Kit (Fermentas). Quantitative PCR was performed through the BioRad Chromo4 real-time PCR system. At the end point of PCR cycles, melt curves were made to check product purity. The mRNA levels of NCOA3 were expressed as a ratio relative to the GAPDH mRNA in each sample. Exploratory data analysis using box plot was applied to visually identify the expression level of target mRNA.
Statistical analysis was performed with the SPSS Software (version 12). Data were analyzed by the χ2 test or Fisher exact test. P values less than 0.05 were considered statistically significant. Results of the NCOA3 mRNA expression in normal and tumor tissue samples were compared using two-way repeated measurement ANOVA. One-way, repeated measurement analysis of variance (ANOVA-RM) was performed at a significance level of P = 0.05 to determine differences from controls within each group. Two-way analysis of variance (ANOVA-2) was performed after baseline subtraction, at a significance level of P = 0.05, to determine differences between the groups with amplified and unaltered NCOA3 copy number.
Since no statistical difference of CNVs between ANTs and healthy normal controls was observed, the adjacent normal tissues could be used as controls for the cancer tissues in this study (Table 1). Table 2 shows CNVs of NCOA3 in paired samples of CRCs and ANTs. A total of 142 CRC samples were examined. Thirty-nine (27.5%) of CRC samples showed gains of NCOA3. Less than 20% of the CRC tissue samples from patients with early-stage CRC (Dukes A and B) had either three, four, or more than four copies of the NCOA3 gene, whereas more than 20% of the samples from patients with advanced (Dukes C and D) had either three, four, or more than four copies of NCOA3. There was a correlation between gene copy number gains and the tumor phenotypes (P = 0.0112).
| Samples | Copy number | ||||||
| n | Deletion | Amplification | P valuevs HNC | ||||
| 0 | 1 | 2 | 3 | > 3 | |||
| HNC | 152 | 2 | 8 | 135 | 6 | 1 | |
| ANT | 142 | 3 | 7 | 124 | 5 | 3 | 0.776 |
| Copy numbers | |||||
| CNVs, population | Amplification | P value (vs ANT) | P value (vs Dukes A and B) | ||
| n | ≤2 | >2 | |||
| Total | |||||
| ANT | 142 | 134 | 8 | - | - |
| CRC | 103 | 39 | < 0.0001 | - | |
| Duke’s A and B | |||||
| ANT | 79 | 74 | 5 | - | - |
| CRC | 64 | 15 | 0.0167 | - | |
| Duke’s C and D | |||||
| ANT | 63 | 60 | 3 | - | - |
| CRC | 39 | 24 | < 0.0001 | 0.0112 | |
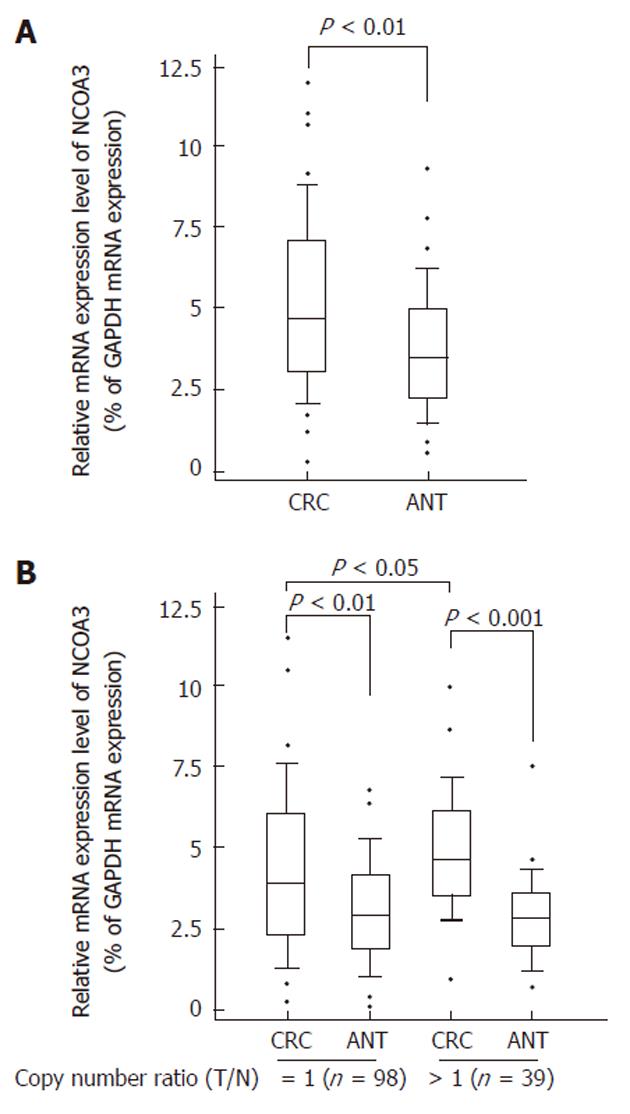
To find whether CNVs of NCOA3 have genotype-phenotype correlation, we compared the mRNA expression levels of NCOA3 between the CRC samples and matched adjacent normal tissues by quantitative real-time RT-PCR. As shown in Figure 1A, increased mRNA expression level of NCOA3 was observed in collected CRC tissues, which was consistent with some previous findings[6,18].
Gene CNVs can contribute to qualitative and quantitative diversities to their gene products. Next, we selected the samples with increased or unaltered copies of NCOA3 and tested whether the NCOA3 mRNA expression was correlated with the copy numbers. The samples with decreased copies of NCOA3 (5 of 142) were not included due to the small sample size. As shown in Figure 1B, the CRC samples in the group with increased or unaltered copies of NCOA3 both showed an increased expression of mRNA compared with adjacent normal tissues (P < 0.01). There was a significant statistical difference between the CRC samples in the groups with increased and unaltered copies of NCOA3 (P = 0.0023). Thus the DNA copy gain at least plays a partial role in the over-expression of NCOA3 in CRCs.
Although cancer is mostly regarded as an acquired disease, familial predisposition plays a significant role in many cancer types. Several highly aggressive cancer predisposing genes correlated with CRC have been identified (DNA mismatch repair genes[19], APC, SMAD4, BMPR1A and MUTYH[20]). As yet, however, these genes explain only a fraction of the familial and/or hereditary cases of cancer. CNVs identified by CGH and array technology have clearly been shown to have the potential to directly or indirectly influence a healthy individual’s susceptibility to cancer, for example by varying the gene dosage of tumor suppressors or oncogenes[21,22]. However, there are many discrepancies among previous studies which used high-resolution approaches to screen CNVs[23-27]. Thus, validation of such CNVs by a larger amount of clinical samples is required. In the present study, CNVs of NCOA3 were examined in a relatively high number of case-matched CRC samples. The results obtained here were more definitive than previous reports.
Examination of the CNVs of oncogenes or tumor suppressor genes is a starting point for investigations into the role of gene amplification in the colorectal carcinogenic process. In this study, no differences of NCOA3 gene copy number were found between the healthy normal controls and ANTs from CRC patients. Thus the CNVs of NCOA3 in CRCs were more likely acquired DNA aberrations. The frequency of gains of NACO3 in CRCs was consistent with previous studies, where gains of chromosomes 20q12 were reported in 20%-60% of CRCs, respectively[11,12]. There was a correlation between the frequency of DNA copy-number gains of NCOA3 and Dukes classifications of the CRC samples (P = 0.0112), which suggested the potential role of altered copy number of NCOA3 gene in the progression of CRCs.
It is expected that the CNVs do have a genotype-phenotype correlation. Phenotypic effects of genetic differences, such as CNVs, are supposedly brought about by changes in expression levels[28,29]. We investigated the correlation between the NCOA3 mRNA expression and the copy numbers of its DNA. Contrary to our expectation, the correlation was not as positive as predicted, although a statistical difference was found. mRNA expression of NCOA3 was increased in both the groups of increased and unchanged DNA copies. There was a statistical difference of mRNA expression between the groups of increased and unaltered DNA copies (P < 0.05). Thus CNVs did play a role of over-expression of the NCOA3 mRNA in CRCs while there were also other mechanisms involved. This is consistent with two recent reports which assessed an over-representation of differentially expressed genes among CNV-mapping transcripts, showing a weak yet significant positive correlation between relative expression level and gene dosage[30,31].
In a small percentage of samples, the recorded relative expression levels were inversely correlated with copy numbers (data not shown here). Such kinds of inverse correlation were also observed by other previous studies[30,31]. The mechanism of this phenomenon is still poorly understood but may be explained by two models. In the first model, a negative correlation between the number of copies and relative expression is explained by immediate early genes, which induced the expression of a repressor directly or indirectly. This expression, by a negative feedback loop, reduces or even abolishes the expression of the CNV gene. In the second model, the extra copies of a gene impair, through steric hinderance, their access to a specific transcription factory, where this particular locus should be transcribed.
Taken together, our findings showed that the CNVs of NCOA3 had a correlation with CRC progression as well as gene expression and might have the potential to serve as a prognostic marker for colorectal malignancies. However, the functional consequences of CNVs, the different features of CNVs between colorectal and other gastrointestinal malignancies and the underlying mechanisms of the heterogeneous expression levels, need to be extensively investigated in the future.
We thank Shenzhen Biomedical Research Support Platform for the technical help.
NCOA3 is a member of the p160 nuclear receptor co-activator family and is considered an important oncogene. The chromosome locus of NCOA3 gene, 20q12, is frequently amplified in colorectal cancers (CRCs) and the functional impact of such regions needs to be extensively investigated in a large size of clinical samples.
Fluorescence in situ hybridization and array-CGH are the regular methods for the detection of genomic imbalances in tumors. However, the obtained data from these approaches are generally informative but not definitive. Recently, studies using quantitative real-time polymerase chain reaction can provide more precise data on the analysis of gene copy number.
To date, there has been a limited number of studies on the copy number variation (CNV) of NCOA3 gene in tumors. The present study employed a more sensitive method to investigate the copy number variation of the targeted gene in a larger number of CRC samples. Furthermore, the authors elucidated the potential correlation between gene copy number and tumor progression.
This study indicated the different mechanisms of NCOA3 regulation in CRCs. This may impact the efficacy of NCOA3 targeted therapies.
CNVs are alterations of the DNA of a genome that results in the cell having an abnormal number of copies of one or more sections of the DNA. CNVs correspond to relatively large regions of the genome that have been deleted (fewer than the normal number) or duplicated (more than the normal number) on certain chromosomes.
The authors investigated the role NCOA3 gene in CRC. Authors detected copy number gain of NCOA3 in a relatively high percentage of CRC samples and correlated them with Dukes stage of CRC. Moreover, they observed a positive correlation between CNV gain and mRNA levels of NCOA3 gene. The study is original and potentially interesting.
Peer reviewer: Ondrej Slaby, PhD, Assistant Professor, Masaryk Memorial Cancer Institute, Department of Comprehensive Cancer Care, Zluty kopec 7, 65653 Brno, Czech Republic
S- Editor Tian L L- Editor Ma JY E- Editor Zhang DN
| 1. | Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615-630. [PubMed] [DOI] [Full Text] |
| 2. | Gojis O, Rudraraju B, Alifrangis C, Krell J, Libalova P, Palmieri C. The role of steroid receptor coactivator-3 (SRC-3) in human malignant disease. Eur J Surg Oncol. 2010;36:224-229. [PubMed] |
| 3. | Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin. 2006;27:387-394. [PubMed] [DOI] [Full Text] |
| 4. | Xu Y, Chen Q, Li W, Su X, Chen T, Liu Y, Zhao Y, Yu C. Overexpression of transcriptional coactivator AIB1 promotes hepatocellular carcinoma progression by enhancing cell proliferation and invasiveness. Oncogene. 2010;29:3386-3397. [PubMed] [DOI] [Full Text] |
| 5. | He LR, Liu MZ, Li BK, Rao HL, Deng HX, Guan XY, Zeng YX, Xie D. Overexpression of AIB1 predicts resistance to chemoradiotherapy and poor prognosis in patients with primary esophageal squamous cell carcinoma. Cancer Sci. 2009;100:1591-1596. [PubMed] [DOI] [Full Text] |
| 6. | Xie D, Sham JS, Zeng WF, Lin HL, Bi J, Che LH, Hu L, Zeng YX, Guan XY. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol. 2005;36:777-783. [PubMed] [DOI] [Full Text] |
| 7. | Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78-85. [PubMed] |
| 8. | Andrews J, Kennette W, Pilon J, Hodgson A, Tuck AB, Chambers AF, Rodenhiser DI. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One. 2010;5:e8665. [PubMed] [DOI] [Full Text] |
| 9. | Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mossé YP, Wood A, Lynch JE. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987-991. [PubMed] [DOI] [Full Text] |
| 10. | Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, Sun J, Wiklund F, Wiley K, Isaacs SD. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69:2176-2179. [PubMed] [DOI] [Full Text] |
| 11. | Lukásová E, Kozubek S, Falk M, Kozubek M, Zaloudík J, Vagunda V, Pavlovský Z. Topography of genetic loci in the nuclei of cells of colorectal carcinoma and adjacent tissue of colonic epithelium. Chromosoma. 2004;112:221-230. [PubMed] [DOI] [Full Text] |
| 12. | Horoyan M, Benoliel AM, Capo C, Bongrand P. Localization of calcium and microfilament changes in mechanically stressed cells. Cell Biophys. 1990;17:243-256. [PubMed] |
| 13. | Nicolet C, Guérin E, Neuville A, Kerckaert JP, Wicker N, Bergmann E, Brigand C, Kedinger M, Gaub MP, Guenot D. Evidence for various 20q status using allelotyping, CGH arrays, and quantitative PCR in distal CIN colon cancers. Cancer Lett. 2009;282:195-204. [PubMed] [DOI] [Full Text] |
| 14. | Yu B, Shao Y, Li P, Zhang J, Zhong Q, Yang H, Hu X, Chen B, Peng X, Wu Q. Copy number variations of the human histamine H4 receptor gene are associated with systemic lupus erythematosus. Br J Dermatol. 2010;163:935-940. [PubMed] [DOI] [Full Text] |
| 15. | Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444-454. [PubMed] [DOI] [Full Text] |
| 16. | Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, Tothill R, Okamoto A, Raeder MB, Harnett P. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417-1427. [PubMed] [DOI] [Full Text] |
| 17. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Full Text] |
| 18. | Grivas PD, Tzelepi V, Sotiropoulou-Bonikou G, Kefalopoulou Z, Papavassiliou AG, Kalofonos H. Estrogen receptor alpha/beta, AIB1, and TIF2 in colorectal carcinogenesis: do coregulators have prognostic significance? Int J Colorectal Dis. 2009;24:613-622. [PubMed] [DOI] [Full Text] |
| 19. | Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, Campbell H, Dunlop MG. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751-2763. [PubMed] [DOI] [Full Text] |
| 20. | Tenesa A, Campbell H, Barnetson R, Porteous M, Dunlop M, Farrington SM. Association of MUTYH and colorectal cancer. Br J Cancer. 2006;95:239-242. [PubMed] [DOI] [Full Text] |
| 21. | Dear PH. Copy-number variation: the end of the human genome? Trends Biotechnol. 2009;27:448-454. [PubMed] [DOI] [Full Text] |
| 22. | Shlien A, Tabori U, Marshall CR, Pienkowska M, Feuk L, Novokmet A, Nanda S, Druker H, Scherer SW, Malkin D. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2008;105:11264-11269. [PubMed] [DOI] [Full Text] |
| 23. | Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, Yamrom B, Alex G, Pai D, Navin N. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood. 2009;113:1294-1303. [PubMed] [DOI] [Full Text] |
| 24. | Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Göransson H, Liljedahl U, Ralfkiaer U, Mansouri M, Buhl AM, Smedby KE. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697-711. [PubMed] [DOI] [Full Text] |
| 25. | Schäfer M, Schwender H, Merk S, Haferlach C, Ickstadt K, Dugas M. Integrated analysis of copy number alterations and gene expression: a bivariate assessment of equally directed abnormalities. Bioinformatics. 2009;25:3228-3235. [PubMed] [DOI] [Full Text] |
| 26. | Strefford JC, van Delft FW, Robinson HM, Worley H, Yiannikouris O, Selzer R, Richmond T, Hann I, Bellotti T, Raghavan M. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci USA. 2006;103:8167-8172. [PubMed] [DOI] [Full Text] |
| 27. | Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, Minto L, Barber KE, Parker H, Wright SL. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100-107. [PubMed] [DOI] [Full Text] |
| 28. | Dermitzakis ET, Stranger BE. Genetic variation in human gene expression. Mamm Genome. 2006;17:503-508. [PubMed] [DOI] [Full Text] |
| 29. | Reymond A, Henrichsen CN, Harewood L, Merla G. Side effects of genome structural changes. Curr Opin Genet Dev. 2007;17:381-386. [PubMed] [DOI] [Full Text] |