Published online Dec 28, 2012. doi: 10.3748/wjg.v18.i48.7319
Revised: November 5, 2012
Accepted: November 11, 2012
Published online: December 28, 2012
Processing time: 138 Days and 18.3 Hours
AIM: To elucidate high mobility group-box 3 (HMGB3) protein expression in gastric adenocarcinoma, its potential prognostic relevance, and possible mechanism of action.
METHODS: Ninety-two patients with gastric adenocarcinomas surgically removed entered the study. HMGB3 expression was determined by immunohistochemistry through a tissue microarray procedure. The clinicopathologic characteristics of all patients were recorded, and regular follow-up was made for all patients. The inter-relationship of HMGB3 expression with histological and clinical factors was analyzed using nonparametric tests. Survival analysis was carried out by Kaplan-Meier (log-rank) and multivariate Cox (Forward LR) analyses between the group with overexpression of HMGB3 and the group with low or no HMGB3 expression to determine the prognosis value of HMGB3 expression on overall survival. Further, HMGB3 expression was knocked down by small hairpin RNAs (shRNAs) in the human gastric cancer cell line BGC823 to observe its influence on cell biological characteristics. The MTT method was utilized to detect gastric cancer cell proliferation changes, and cell cycle distribution was analyzed by flow cytometry.
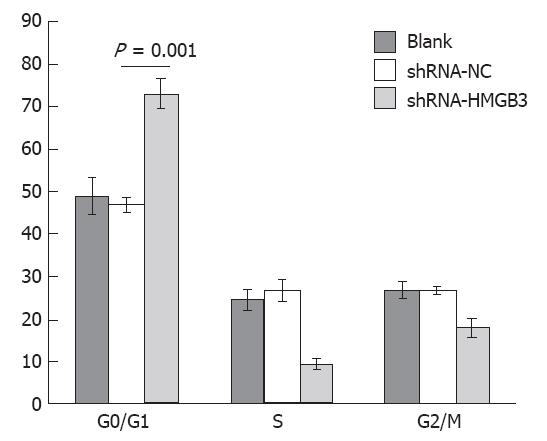
RESULTS: Among 92 patients with gastric adenocarcinomas surgically removed in this study, high HMGB3 protein expression was detected in the gastric adenocarcinoma tissues vs peritumoral tissues (P < 0.001). Further correlation analysis with patients’ clinical and histology variables revealed that HMGB3 overexpression was obviously associated with extensive wall penetration (P = 0.005), a positive nodal status (P = 0.004), and advanced tumor-node-metastasis (TNM) stage (P = 0.001). But there was no correlation between HMGB3 overexpression and the age and gender of the patient, tumor localization or histologic grade. Statistical Kaplan-Meier survival analysis disclosed significant differences in overall survival between the HMGB3 overexpression group and the HMGB3 no or low expression group (P = 0.006). The expected overall survival time was 31.00 ± 3.773 mo (95%CI = 23.605-38.395) for patients with HMGB3 overexpression and 49.074 ± 3.648 mo (95%CI = 41.925-57.311) for patients with HMGB3 no and low-level expression. Additionally, older age (P = 0.040), extensive wall penetration (P = 0.008), positive lymph node metastasis (P = 0.005), and advanced TNM tumor stage (P = 0.007) showed negative correlation with overall survival. Multivariate Cox regression analysis indicated that HMGB3 overexpression was an independent variable with respect to age, gender, histologic grade, extent of wall penetration, lymph nodal metastasis, and TNM stage for patients with resectable gastric adenocarcinomas with poor prognosis (hazard ratio = 2.791, 95%CI = 1.233-6.319, P = 0.019). In the gene function study, after HMGB3 was knocked down in the gastric cell line BGC823 by shRNA, the cell proliferation rate was reduced at 24 h, 48 h and 72 h. Compared to BGC823 shRNA-negative control (NC) cells, the cell proliferation rate in cells that had HMGB3 shRNA transfected was significantly decreased (P < 0.01). Finally, cell cycle analysis by FACS showed that BGC823 cells that had HMGB3 knocked down were blocked in G1/G0 phase. The percentage of cells in G1/G0 phase in BGC823 cells with shRNA-NC and with shRNA-HMGB3 was 46.84% ± 1.7%, and 73.03% ± 3.51% respectively (P = 0.001), whereas G2/M cells percentage decreased from 26.51% ± 0.83% to 17.8% ± 2.26%.
CONCLUSION: HMGB3 is likely to be a useful prognostic marker involved in gastric cancer disease onset and progression by regulating the cell cycle.
- Citation: Tang HR, Luo XQ, Xu G, Wang Y, Feng ZJ, Xu H, Shi YW, Zhang Q, Wu LG, Xue CQ, Wang CW, Wu CY. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol 2012; 18(48): 7319-7326
- URL: https://www.wjgnet.com/1007-9327/full/v18/i48/7319.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i48.7319
Gastric cancer is reported to be one of the common deva-stating types of cancer. It has a high occurrence rate, short survival period and high mortality rate. The mortality rate can be up to 25.2/100 000 in China, which accounts for 25% of deaths in malignant cancer. The occurrence ratio and mortality are ranked as No. 1 and No. 2 in malignant cancer[1]. Usually gastric cancer is already at a late stage when it is diagnosed; the 5-year survival rate in clinical stage III and IV patients is only about 28.4% and 7.6%, respectively. There are many factors including lymph node metastasis, extent of wall penetration, pathological tumor-node-metastasis (pTNM) stage, and surgical mode etc., associated with prognosis of the disease. Those factors are more clinipathologically related, however, investigating molecular biomarkers can not only provide indications for clinic prognosis, but also can identify potential targets for clinical therapy.
High mobility group-box 3 (HMGB3) belongs to the high mobility group box (HMG-box) subfamily. It has 99% homology with HMGB1 and HMGB2[2]. This subfamily has a HMG-box, so it can regulate gene transcription by participating with formation of enhancesomes. The HMG-box subfamily also plays an important role in DNA replication, transcription, recombination and repair, etc.[3,4]. HMGB3 was reported to have high expression in embryos and weak expression in adult tissues. HMGB3 is important in keeping the balance between self-renewal and differentiation status in mouse hematopoietic stem cells[5]. Also it was known that HMGB3 is indispensible in maintaining mouse leukemia stem cell self-renewal capability. Simultaneously overexpression of HMGB3, c-MyB and CBX5 can make hematopoietic stem/progenitor cells immortalized[6]. Recent studies showed that HMGB3 and NPU98 fusion protein forms a new oncogenic gene in leukemia[7]. HMGB3 also participates in recurrence of acute lymphoid leukemia and it shows high expression in the progression phase of breast cancer[8]. HMGB3 was identified as one of the bio-markers detected in peripheral blood in lung cancer[9]. Though HMGB3 is found in some types of cancers, its role in gastric cancer is still unclear.
In this study, we observed the expression of HMGB3 in gastric adenocarcinoma tissues by immunochemistry, and analyzed its correlation between expression level and clinicopathologic variables and prognosis. We found that HMGB3 showed a high expression level in gastric cancer vs peritumoral tissues. And HMGB3 overexprssion was obviously associated with extensive wall penetration, a positive nodal status, advanced TNM stage and poor prognosis. Moreover, we silenced HMGB3 expression in BGC823 gastric cancer cell line by small interfering RNA (siRNA), and observed the changes in cell proliferation and cell cycle. BGC823 cells with HMGB3 knocked down showed a decreased proliferation rate and the ratio of G0/G1 phase cells in the cell cycle significantly increased. Results above indicate that the overexpression of HMGB3 is a marker for poor prognosis of gastric adenocarcinomas and that it may function by affecting gastric cell proliferation and cell cycle.
Tissue microarrays (TMAs) from a total of 92 consecutive cases of gastric adenocarcinomas operated on in our hospital from December 2006 to October 2007 were prepared for immunohistochemical testing. All the patients were given radical resection and D1+ or D2 lymphadenectomy followed by adjuvant chemotherapy with the regimen epirubicin, cisplatin and fluorouracil. No preoperative therapy was given to any patient. The pathologic staging was made according to American Joint Committee on Cancer TNM staging system. The follow up end point was defined as the death of patients. The use of the tissue samples in TMA analysis and clinical data was approved by Medical Ethics Committee of Jiangsu University and the patients. Patients’ clinical and histopathologic data were summarized in Table 1.
| Variables | Samples | HMGB3 overexpression | P value |
| Age at surgery (yr) | |||
| ≤ 60 | 34 (36.96) | 20 (60.00) | 0.247 |
| > 60 | 58 (63.04) | 30 (51.16) | |
| Gender | |||
| Male | 61 (66.30) | 30 (50.00) | 0.649 |
| Female | 31 (33.70) | 20 (62.5) | |
| Extent of wall penetration | |||
| pT1-2 | 22 (23.91) | 6 (26.67) | 0.005 |
| pT3-4 | 70 (76.01) | 44 (63.46) | |
| Lymph node metastasis | |||
| N0 | 27 (29.3) | 5 (14.29) | 0.004 |
| N1-3 | 65 (70.65) | 45 (70.59) | |
| Tumor stage | |||
| StageI+ II | 37 (40.22) | 9 (23.21) | 0.001 |
| Stage III + IV | 55 (59.78) | 41 (56.76) | |
| Tumor localisation | |||
| Fundus gastricus and cardia | 11 (11.96) | 6 (54.55) | 0.955 |
| Gastric body | 33 (35.87) | 21 (63.64) | |
| Gastric antrum | 48 (52.17) | 23 (47.91) | |
| Histologic grade | |||
| G1 + 2 | 22 (23.91) | 9 (40.91) | 0.073 |
| G3 + 4 | 70 (76.09) | 41 (58.57) |
For each case, we selected the tumor foci for the TMA construction during routine diagnosis by marking them on the more representative hematoxylin-eosin-stained slide with a waterproof pencil. At the same time we chose corresponding peritumoral tissue as a control. The advanced tissue arrayer (ATA-100, Chemicon International, Tamecula, CA, United States) was used to create holes in a recipient paraffin block to acquire cylindrical core tissue biopsies with a diameter of 1 mm from the specific areas of the “donor” block. The tissue core biopsies were transferred to the recipient paraffin block at defined array positions. The TMAs contained tissue samples from 92 formalin-fixed, paraffin-embedded cancer specimens with known diagnosis, and correlated noncancerous tissues from the same patients. The block was incubated in an oven at 45 °C for 20 min to allow complete embedding of the grafted tissue cylinders in the paraffin of the recipient block, and then stored at 4 °C until microtome sectioning.
Rabbit-derived anti-human HMGB3 antibody (Epitomics, Cat. #2416-1) were used for immunohistochemical (IHC) detection of HMGB3 protein in TMAs. TMA sections were processed for IHC demonstration of HMGB3 protein by the Biotin-Avidin-Peroxidase detection system (Sigma). The anti-HMGB3 antibodies were used at 1:50 dilutions. Endogenous peroxidase was inhibited by incubation with freshly prepared 3% hydrogen peroxide with 0.1% sodium azide. Nonspecific staining was blocked with 0.5% casein and 5% normal goat serum. TMAs were incubated with biotinylated goat anti-rabbit antibodies and ExtrAvidin-conjugated horseradish peroxidase. Staining was developed with diaminobenzidine substrate and sections were counterstained with hematoxylin. Normal mouse serum or phosphate-buffered saline (PBS) replaced anti-HMGB3 antibodies used as negative controls.
HMGB3 expression was semiquantitatively estimated as the total HMGB3 immunostaining score, which was calculated as the sum of a proportion score and an intensity score. The proportion score reflects the fraction of positive staining cells (score 0, < 5%; score 1, 5%-10%; score 2, 10%-50%; score 3, 50%-75%; score 4, > 75%). The intensity score represents the staining intensity (score 0, no staining signal; score 1, weak positive signal; score 2, moderate positive signal; score 3, strong positive signal). Finally, a total expression score was given ranging from 0 to 12. Based on the analysis in advance, the overexpression of HMGB3 was defined as a total expression score ≥ 9[10].
The human gastric cancer cell line BGC823 was obtained from the American Type Culture Collection (Rockville, MD). The cells were cultured in Dulbecco’s modified Eagle’s minimal essential medium media supplemented with 10% fetal bovine serum (Gibco), penicillin (100 IU/mL) and streptomycin (100 mg/mL) and grown in a humidified incubator with a 5% CO2 atmosphere at 37 °C.
Human HMGB3 small hairpin RNA (shRNA) and control shRNA were obtained from Shanghai R and S Biotechnology Co., Ltd, and were transfected into cells using Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA, United States) according to the manufacturer’s instructions.
Cells were plated at a density of 5 × 103 cells/well in 96-well plates in 200 mL medium. After siRNA was tranfected for 24 h, 48 h and 72 h, 20 μL MTT was added to each well. The mixture was incubated at 37 °C for 4 h in the dark, washed to remove media and MTT, and 200 μL dimethyl sulfoxide added to each well. The absorption value at 490 nm was recorded.
The BGC823/blank, BGC823/small hairpin RNAs-negative control (shRNAs-NC) shRNA-NC and BGC823/shRNA-HMGB3 cells were seeded in six-well plates at a concentration of 5 × 105 cells/mL. Cells were harvested following trypsinization, washed once with cold PBS and fixed in 70% ethanol for 24 h. DNA staining was carried out by resuspending the cells in a solution of PBS containing 20 mg/mL of RNase A, 50 mg/mL of propidium iodide, with subsequent incubation at 37 °C for 30 min. The stained cells were analysed for the FL-2 area using a flow cytometer (Beckman Coulter, Brea, CA), and DNA histograms were analyzed using Modifit software. Experiments were performed in triplicate. Results are presented as percentage of cells in a particular phase.
Data was analyzed using the statistical package for the Social Sciences Version 16.0 (SPSS 16.0). For clinical data statistical analysis, the inter-relationship of HMGB3 expression with histology or clinical factors was analyzed using nonparametric tests. The screen of significant factors associated with survival rate in patients with gastric adenocarcinoma used Kaplan-Meier and log-rank test; Cox regression analysis (Forward LR) was used for multivariate analysis and to determine the 95% confidence interval. Cell experiment data were expressed as mean ± SD of three independent experiments. Significance of differences between groups was determined by one-way analysis of variance. The significance was set at P < 0.05.
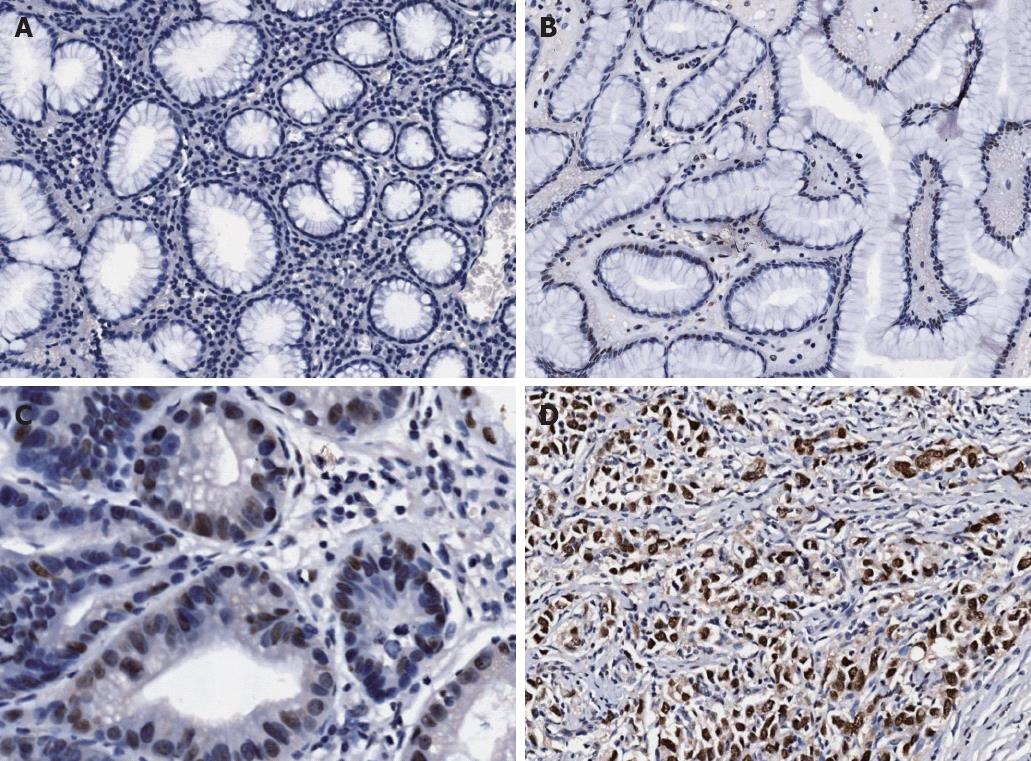
Expression of HMGB3 protein was evaluated by immunohistochemical staining. In gastric adenocarcinoma cells, the expression of HMGB3 protein was mainly found in the nucleus, and weakly detected in cytoplasm (Figure 1). Positive staining was detected at a level of 94.57% (87/92) in gastric adenocarcinoma tissue, and 52.17% (48/92) in peritumoral tissue with significant difference (P < 0.001). The rate of HMGB3 overexpression (total expression score ≥ 9) was elevated in gastric adenocarcinoma tissue compared with corresponding peritumoral tissue (54.35% vs 0.00%, P < 0.001). The difference in HMGB3 expression between peritumoral and normal (distant) tissues was not assessed. We also detected the HMGB3 mRNA level in gastric adenocarcinoma tissue and the corresponding peritumoral tissues by using quantitative reverse transcriptase-polymerase chain reaction, and it was found that HMGB3 mRNA expression level was significantly higher in gastric cancer than in peritumoral samples (detailed data not shown).
We analyzed the correlation between HMGB3 expression and age, gender, tumor localization, histologic grade, extent of wall penetration, lymph node metastasis and tumor stage. The results showed that HMGB3 expression level was much higher in the pT3-4 group compared to the pT1-2 group (P = 0.005); it was also much higher in the N1-3 group compared to the N0 group (P = 0.004) and in the stage III + IV group compared to the stageI+ II group (P = 0.001); all the results were statistically significant. But the correlation between HMGB3 expression and the age and gender of the patient and tumor localization, histologic grade had no statistical significance (Table 1).
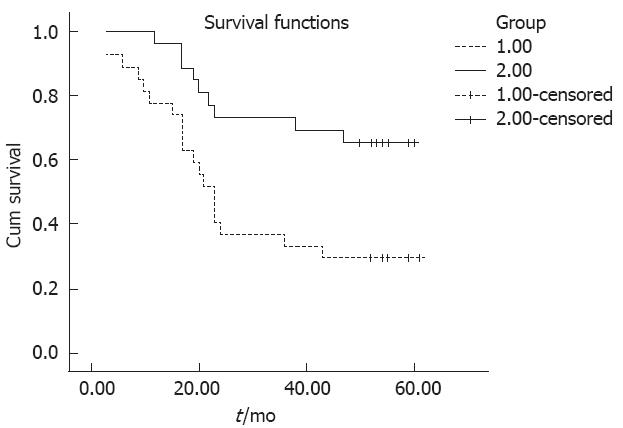
We used Kaplan-Meier survival analysis to analyze the gender, age, tumor localization, histologic grade, extent of wall penetration, lymph node metastasis, tumor stage and HMGB3 expression correlation with patient’s prognosis. It was observed that there was no correlation between gender, tumor localization and histologic grade with prognosis. But age (P = 0.040), extent of wall penetration (P = 0.008), lymph node metastasis (P = 0.005), TNM tumor stage (P = 0.007) and HMGB3 overexpression (P = 0.006) showed a correlation with overall survival (Table 2). This survival analysis revealed that HMGB3 overexpression affected overall survival. There was a significant difference in overall survival between groups with HMGB3 overexpression and with no or low level expression (Table 2 and Figure 2). The expected overall survival time was 31.00 ± 3.773 mo for tumors with HMGB3 overexpression (95%CI = 23.605-38.395) and 49.074 ± 3.648 mo for tumors with HMGB3 no or low level expression (95%CI = 41.925-57.311). Multivariate Cox regression analysis indicate that HMGB3 overexpression was predictive of mortality (hazard ratio = 2.791, 95%CI = 1.233-6.319, P = 0.019), and as an independent variable with respect to age, gender, histologic grade, extent of wall penetration, lymph node metastasis, and TNM stage for patients with resectable gastric adenocarcinomas.
| Clinicopathological characteristics | Samples (n) | Average survival (mo) | 95%CI | P value |
| Sex | ||||
| Male | 61 | 41.563 ± 2.823 | 36.031-47.096 | 0.308 |
| Female | 31 | 37.24 ± 4.027 | 29.347-45.133 | |
| Age (yr) | ||||
| ≤ 60 | 34 | 47.409 ± 4.113 | 39.348-55.470 | 0.040 |
| > 60 | 58 | 35.143 ± 3.797 | 27.700-42.585 | |
| Histologic grade | ||||
| G1 + G2 | 22 | 40.938 ± 5.238 | 30.670-51.205 | 0.323 |
| G3 + G4 | 70 | 37.366 ± 3.366 | 30.769-43.963 | |
| Lymph node status | ||||
| pN0 | 27 | 54.800 ± 1.885 | 51.085-58.515 | 0.005 |
| pN1-3 | 65 | 33.158 ± 3.646 | 26.012-40.304 | |
| Extent of wall penetration | ||||
| pT1 + 2 | 22 | 48.385 ± 3.949 | 40.645-56.124 | |
| pT3 + 4 | 70 | 33.447 ± 3.447 | 26.692-40.203 | 0.008 |
| TNM stage | ||||
| StageI+ II | 37 | 47.511 ± 2.927 | 41.774-53.248 | 0.007 |
| Stage III + IV | 55 | 32.949 ± 3.266 | 26.547-39.352 | |
| HMGB3 expression | ||||
| No or low expression | 42 | 49.074 ± 3.648 | 41.925-57.311 | 0.006 |
| Overexpression | 50 | 31.00 ± 3.773 | 23.605-38.395 | |
| Location | ||||
| Stomach fundus | 11 | 31.455 ± 7.193 | 17.356-45.553 | 0.2261 |
| Gastric body | 33 | 41.395 ± 3.640 | 34.226-48.493 | 0.2972 |
| Gastric antrum | 48 | 41.410 ± 3.211 | 35.116-47.703 | 0.9223 |
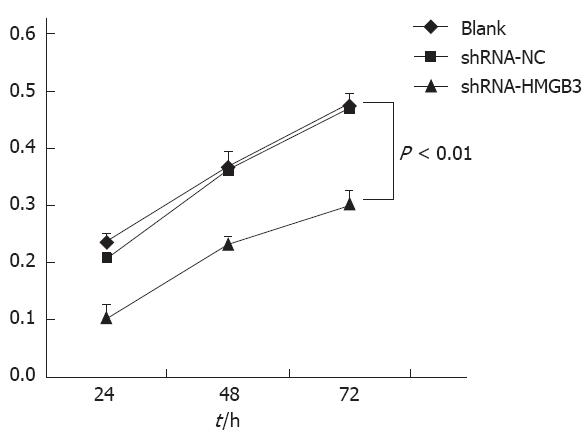
To determine whether HMGB3 RNAi had an inhibitory effect on gastric cancer cell line BGC823 cell proliferation, we measured cell growth with an MTT assay. Data demonstrated that after HMGB3 was silenced, the cell proliferation rate significantly reduced in 24 h, 48 h, 72 h. Compared to BGC823 cells with shRNA-NC, cell proliferation rate in cells that have HMGB3 siRNA transfected was decreased significantly (P < 0.01; Figure 3). These results indicate that knockdown of HMGB3 inhibits cell proliferation.
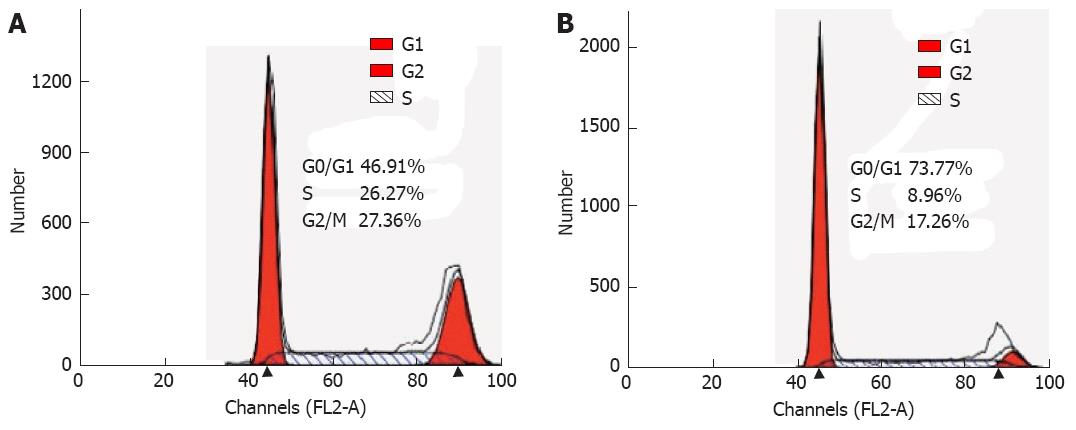
To identify a potential mechanism for HMGB3-specific silencing-mediated reduction of cell proliferation, cell cycle distribution was assessed using flow cytometry. The analysis showed that the percentage of cells in G0/G1 phase increased significantly (P = 0.001), whereas cells in the G2/M phases decreased in BGC823/HMGB3 RNAi cells compared with their parental controls. The percentage of cells in G1/G0 phase in BGC823 cells with shRNA-NC and with shRNA-HMGB3 was 46.84% ± 1.7% and 73.03% ± 3.51% respectively, whereas the percentage of cells in G2/M was 26.51% ± 0.83% and 17.8% ± 2.26%. These data suggest that HMGB3 silencing may induce G0/G1 cell arrest (Figures 4 and 5).
Results above showed that high expression of HMGB3 in gastric cancer positively correlated with extensive wall penetration, positive lymph node metastasis, advanced tumor stage, and poor prognosis. Multivariable Cox regression indicated that this protein overexpression can be used as an independent biomarker in prognosis of gastric cancer. It implies that HMGB3 might participate in gastric cancer onset and development. Further silencing of HMGB3 expression in the gastric cancer cell line by shRNA showed that cell proliferation decreased and the cell cycle was blocked in G0/G1 phase. These results indicate that HMGB3 had a role in the proliferation and cell cycle of gastric cancer cells.
The onset and progression of gastric cancer correlated with various molecular and genetic incidents. To investigate the significance of the molecular expression in gastric cancer may help us to identify potential treatment targets and (or) predictive markers of prognosis.
The HMG Box subfamily is a family of non-histone chromosomal proteins; these members including HMGB1, HMGB2 and HMGB3[2]. HMGB family is thought to play a fundamental role in DNA replication, nucleosome assembly and transcription. There are studies overwhelmingly focusing on HMGB1, since it is widely expressed in all kinds of cell types in adult vertebrates. HMGB1 has been shown to interact with recombination activating gene 1 (RAG1) and RAG2 to play a role in immunology and inflammation, and is associated with proliferation and metastasis of many tumor types[11,12]. However, little is known about the function of HMGB3.
In this study, we analyzed the role of HMGB3 in gastric cancer. Firstly, we observed HMGB3 expression in gastric adenocarcinoma and found it has a high expression level. The positive expression rate in gastric adenocarcinoma is 94.57% (87/92); the overexpression rate reached 54.35% (50/92). While the positive expression rate in gastric peritumoral tissue is 52.17 % (48/92), no overexpression was found in it. Pourhoseingholi et al[13] reported that HMGB3 also has high expression in progressive breast cancer. A combination of proofs that there is high expression of HMGB3 in recursive leukemia[8] and the pivotal role of HMGB3 in maintaining the self-renewal capability of leukemia stem cells[6] imply that HMGB3 is a critical gene participating in cancer progression. Secondly, we further analyzed the correlation of HMGB3 expression with gastric cancer clinicopathologic variables and prognosis. We found high HMGB3 expression correlates with extensive wall penetration, positive lymph node metastasis and advanced tumor stage, which are the important prognostic factors in gastric cancer. Kaplan-Meier survival analysis showed HMGB3 high expression is negatively correlated with the overall survival of patients with resected gastric adenocarcinoma. The expected overall survival time in high HMGB3 expression patients is 31.000 ± 3.773 mo, while in no or low HMGB3 expression patients it is 49.074 ± 3.648 mo. Multivariate analysis shows HMGB3 overexpression can work as an independent variable for poor survival in resectable gastric adenocarcinoma. In analyzing factors affecting gastric cancer prognosis, different research groups reached more or less different conclusions since the methods used and the samples they analyzed were different. However, there are common factors including age, tumor late stage, small surgical area, large cancer volume, adjuvant chemotherapy etc. correlated with poor prognosis in gastric cancer[14-17]. Many researchers reported that pathological classification is also an important factor affecting prognosis[14,15,17], though in this study it showed a negative result. This could be due to the number of level II and III patients being large, thus affecting the conclusion. Now the diagnosis and therapy of tumors is entering the “molecular moment”. Molecular biomarkers not only could be prognostic factors, but could also be potential therapeutic targets for clinical therapy. Mitogen-activated protein kinase, Janus kinase/signal transducers and activators of transcription and nuclear factor-kappaB were reported to have high expression levels in gastric cancer[18-20]; however, none of them can be used as a prognostic biomarker alone. Ki67 and PNCA have been widely used to indicate a high cancer cell proliferation rate, and both of them are highly expressed in gastric cancer[21]; however, neither has been used to determine prognosis[22,23]. Her2, a therapy target in breast cancer[24,25], is also found in gastric cancer as a new prognostic factor and a novel therapeutic target[26,27]. Lastly, we used the gastric cancer cell line BGC823 to investigate cell proliferation and cell cycle changes after silencing of HMGB3. It was observed that the cell proliferation rate was greatly reduced, and the cell cycle was blocked in G1/G0 phase. This indicates that HMGB3 may promote cell proliferation through cell cycle progression. Researchers found that the number of G2/M cells was significantly reduced in HMGB3-/- mouse hematopoietic progenitor cells and this could be attributed to blockage of the cell cycle in G0/G1 phase[5]. In Xenopus, Terada et al[28] observed that the cell proliferation rate of retinal progenitor cells can be promoted with overexpression of HMGB3. They further discovered that this is accompanied by P27 downregulation[29]. P27 is a member of the cyclin dependent kinase inhibitor family, CIP/KIP, and can prevent activation of cyclin E-CDK6 and cyclin D-CDK4, which consequently blocks cells in G1 phase[30]. Thus, we propose that the mechanism under which HMGB3 promotes cancer onset and development is mainly by enhancing cell proliferation.
In summary, high-level expression of HMGB3 protein was detected in gastric adenocarcinoma cells. The overexpression of HMGB3 was correlated with a poor prognosis of gastric cancer patients. HMGB3 may promote gastric cancer cell proliferation by regulating the cell cycle. Therefore, our data encourages further investigations to elucidate the role of HMGB3 and its molecular mechanism.
Gastric cancer is reported to be one of the common devastating types of cancer that has a high occurrence rate, short survival period and high mortality rate. The mortality rate can be up to 25.2/100 000 in China, which accounts for 25% of deaths in malignant cancer. The occurrence ratio and mortality are ranked as No. 1 and No. 2 in malignant cancer.
Recent studies showed that high mobility group-box3 (HMGB3) and NPU98 fusion protein form a new oncogenic gene in leukemia. HMGB3 also participates in recurrence of acute lymphoid leukemia and it shows a high expression level in the progression phase of breast cancer. HMGB3 was identified as one of the biomarkers detected in peripheral blood in lung cancer. Though HMGB3 is found in some types of cancers, its role in gastric cancer is still unclear.
The authors observed the expression of HMGB3 in gastric adenocarcinoma tissues by immunochemistry, and analyzed its correlation between expression level and clinicopathologic variables and prognosis. Results indicated that the overexpression of HMGB3 is a marker for poor prognosis of gastric adenocarcinomas. HMGB3 may function through affecting gastric cell proliferation and cell cycle.
The results are interesting and suggest that HMGB3 is likely to be a useful prognostic marker involved in gastric cancer disease onset and progression by regulating the cell cycle.
Peer reviewer: Yujin Hoshida, MD, PhD, Cancer Program, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, United States
S- Editor Gou SX L- Editor O’Neill M E- Editor Li JY
| 1. | Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17-20. [PubMed] |
| 2. | Vaccari T, Beltrame M, Ferrari S, Bianchi ME. Hmg4, a new member of the Hmg1/2 gene family. Genomics. 1998;49:247-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170-178. [PubMed] |
| 4. | Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 465] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 5. | Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci USA. 2006;103:13783-13788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 301] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 7. | Petit A, Ragu C, Della-Valle V, Mozziconacci MJ, Lafage-Pochitaloff M, Soler G, Schluth C, Radford I, Ottolenghi C, Bernard OA. NUP98-HMGB3: a novel oncogenic fusion. Leukemia. 2010;24:654-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Staal FJ, de Ridder D, Szczepanski T, Schonewille T, van der Linden EC, van Wering ER, van der Velden VH, van Dongen JJ. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia. 2010;24:491-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Bao G, Qiao Q, Zhao H, He X. Prognostic value of HMGB1 overexpression in resectable gastric adenocarcinomas. World J Surg Oncol. 2010;8:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 956] [Cited by in RCA: 1016] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 11. | Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 547] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 12. | Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427-1431. [PubMed] |
| 13. | Pourhoseingholi MA, Moghimi-Dehkordi B, Safaee A, Hajizadeh E, Solhpour A, Zali MR. Prognostic factors in gastric cancer using log-normal censored regression model. Indian J Med Res. 2009;129:262-267. [PubMed] |
| 14. | Wang W, Li YF, Sun XW, Chen YB, Li W, Xu DZ, Guan XX, Huang CY, Zhan YQ, Zhou ZW. Prognosis of 980 patients with gastric cancer after surgical resection. Chin J Cancer. 2010;29:923-930. [PubMed] |
| 15. | Biglarian A, Hajizadeh E, Kazemnejad A, Zayeri F. Determining of prognostic factors in gastric cancer patients using artificial neural networks. Asian Pac J Cancer Prev. 2010;11:533-536. [PubMed] |
| 16. | Calcagno DQ, Guimarães AC, Leal MF, Seabra AD, Khayat AS, Pontes TB, Assumpção PP, De Arruda Cardoso Smith M, Burbano RR. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Res. 2009;29:2479-2483. [PubMed] |
| 17. | Park JM, Ryu WS, Kim JH, Park SS, Kim SJ, Kim CS, Mok YJ. Prognostic factors for advanced gastric cancer: stage-stratified analysis of patients who underwent curative resection. Cancer Res Treat. 2006;38:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | McCubrey JA, Milella M, Tafuri A, Martelli AM, Lunghi P, Bonati A, Cervello M, Lee JT, Steelman LS. Targeting the Raf/MEK/ERK pathway with small-molecule inhibitors. Curr Opin Investig Drugs. 2008;9:614-630. [PubMed] |
| 19. | Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 226] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 20. | Romano S, Mallardo M, Romano MF. FKBP51 and the NF-κB regulatory pathway in cancer. Curr Opin Pharmacol. 2011;11:288-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Czyzewska J, Guzińska-Ustymowicz K, Pryczynicz A, Kemona A, Bandurski R. Immunohistochemical evaluation of Ki-67, PCNA and MCM2 proteins proliferation index (PI) in advanced gastric cancer. Folia Histochem Cytobiol. 2009;47:289-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Lazăr D, Tăban S, Sporea I, Dema A, Cornianu M, Lazăr E, Goldiş A, Vernic C. Ki-67 expression in gastric cancer. Results from a prospective study with long-term follow-up. Rom J Morphol Embryol. 2010;51:655-661. [PubMed] |
| 23. | Mangham DC, Rowlands DC, Newbold KM, Reynolds GM, Fielding JW, Hallissey MT. Expression of proliferating cell nuclear antigen (PCNA) in gastric carcinoma: no evidence for prognostic value. J Clin Pathol. 1994;47:473-474. [PubMed] |
| 24. | Gianni L, Pienkowski T, Im YH, Roman L, Tseng LM, Liu MC, Lluch A, Staroslawska E, de la Haba-Rodriguez J, Im SA. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1412] [Cited by in RCA: 1670] [Article Influence: 119.3] [Reference Citation Analysis (0)] |
| 25. | Chang HR. Trastuzumab-based neoadjuvant therapy in patients with HER2-positive breast cancer. Cancer. 2010;116:2856-2867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 26. | Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol. 2008;19:1523-1529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 871] [Article Influence: 51.2] [Reference Citation Analysis (2)] |
| 27. | Pazo Cid RA, Antón A. Advanced HER2-positive gastric cancer: Current and future targeted therapies. Crit Rev Oncol Hematol. 2012;Sep 25; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Terada K, Kitayama A, Kanamoto T, Ueno N, Furukawa T. Nucleosome regulator Xhmgb3 is required for cell proliferation of the eye and brain as a downstream target of Xenopus rax/Rx1. Dev Biol. 2006;291:398-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 29. | Terada K, Furukawa T. Sumoylation controls retinal progenitor proliferation by repressing cell cycle exit in Xenopus laevis. Dev Biol. 2010;347:180-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 778] [Article Influence: 45.8] [Reference Citation Analysis (0)] |