Published online Nov 14, 2012. doi: 10.3748/wjg.v18.i42.6096
Revised: July 25, 2012
Accepted: July 28, 2012
Published online: November 14, 2012
AIM: To investigate the variability of the main immunodominant motifs of hepatitis B virus (HBV) core gene by ultra-deep-pyrosequencing (UDPS).
METHODS: Four samples (2 genotype A and 2 genotype D) from 4 treatment-naïve patients were assessed for baseline variability. Two additional samples from one patient (patient 4, genotype D) were selected for analysis: one sample corresponded to a 36-mo treatment-free period from baseline and the other to the time of viral breakthrough after 18 mo of lamivudine treatment. The HBV region analyzed covered amino acids 40 to 95 of the core gene, and included the two main epitopic regions, Th50-69 and B74-84. UDPS was carried out in the Genome Sequencer FLX system (454 Life Sciences, Roche). After computer filtering of UDPS data based on a Poisson statistical model, 122 813 sequences were analyzed. The most conserved position detected by UDPS was analyzed by site-directed mutagenesis and evaluated in cell culture.
RESULTS: Positions with highest variability rates were mainly located in the main core epitopes, confirming their role as immune-stimulating regions. In addition, the distribution of variability showed a relationship with HBV genotype. Patient 1 (genotype A) presented the lowest variability rates and patient 2 (genotype A) had 3 codons with variability higher than 1%. Patient 3 and 4 (both genotype D) presented 5 and 8 codons with variability higher than 1%, respectively. The median baseline frequencies showed that genotype A samples had higher variability in epitopic positions than in the other positions analyzed, approaching significance (P = 0.07, sample 1 and P = 0.05, sample 2). In contrast, there were no significant differences in variability between the epitopic and other positions in genotype D cases. Interestingly, patient 1 presented a completely mutated motif from amino acid 64 to 67 (E64LMT67), which is commonly recognized by T helper cells. Additionally, the variability observed in all 4 patients was particularly associated with the E64LMT67 motif. Codons 78 and 79 were highly conserved in all samples, in keeping with their involvement in the interaction between the HBV virion capsid and the surface antigens (HBsAg). Of note, codon 76 was even more conserved than codons 78 and 79, suggesting a possible role in HBsAg interactions or even in hepatitis B e antigen conformation. Sequential analysis of samples from patient 4 (genotype D) illustrated the dynamism of the HBV quasispecies, with strong selection of one minor baseline variant coinciding with a decrease in core variability during the treatment-free and lamivudine-treated period. The drop in variability seemed to result from a “steady state” situation of the HBV quasispecies after selection of the variant with greatest fitness.
CONCLUSION: Host immune pressure seems to be the main cause of HBV core evolution. UDPS analysis is a useful technique for studying viral quasispecies.
- Citation: Homs M, Buti M, Tabernero D, Quer J, Sanchez A, Corral N, Esteban R, Rodriguez-Frias F. Quasispecies dynamics in main core epitopes of hepatitis B virus by ultra-deep-pyrosequencing. World J Gastroenterol 2012; 18(42): 6096-6105
- URL: https://www.wjgnet.com/1007-9327/full/v18/i42/6096.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i42.6096
Hepatitis B virus (HBV) infection is a global health problem. Around 350 million people are chronically affected with this pathogen, which confers a higher risk of developing liver disease, liver cirrhosis, and hepatocellular carcinoma. The course of HBV infection is closely related to the host immune response and genetic factors[1], and disease progression is related to mutations in the HBV core gene[2-4].
HBV core gene codes for two partially collinear proteins, the hepatitis B e antigen (HBeAg) and hepatitis B core antigen (HBcAg). These proteins, together with the surface antigen (HBsAg) are important targets for antiviral immunity, but HBcAg seems to be the most immunogenic[5]. Several epitopes have been identified in the HBV core gene. Among them, two regions play a particularly important immunodominant role: the sequence from amino acid 50 to 69, which immunostimulates CD4+ T-helper lymphocytes (Th50-69)[6] and the sequence from amino acid 74 to 84, which stimulates B lymphocytes (B74-84)[7,8].
During chronic HBV infection, a large number of amino acid substitutions are seen in the core gene, mainly clustered in epitopic regions. These amino acid changes have been associated with viral persistence because of their impact on the host immune response and the natural course of HBV infection[7,9-14]. The largest number of core gene changes is associated with interferon (IFN) therapy[10-12]. The effect of nucleoside/nucleotide analogues on the core gene has been little investigated, although some variability in a minor epitope (Th28-47) was recently reported[11]. In another recent study, entecavir and adefovir were associated with an enhanced immune response[15]. Selection of core gene amino acid changes might result in evasion of HBV from the host immune system, thereby lengthening the life of infected hepatocytes. For this reason, core gene baseline variability in chronic hepatitis B patients might be crucial for understanding the evolution of the viral quasispecies in response to host immune pressure.
Next-generation sequencing technologies enable deep assessment of gene variability and are especially useful to study the dynamics of viral quasispecies[16-21]. Core gene variability can be studied with this technology, specifically, the 454 FLX platform, which analyzes fragments of 250- to 400-bp length. Although this length does not permit complete analysis of the gene, ultra-deep analysis of the main immunodominant regions of the core protein (Th50-69 and B74-84) is possible. The aim of this study was to analyze the variability of these main HBV core epitopes in chronic hepatitis B patients by ultra-deep pyrosequencing (UDPS).
Four chronic hepatitis B patients with complete clinical documentation were selected for the study; baseline characteristics are indicated in Table 1. All patients were diagnosed with active HBV replication and treated with lamivudine (LVD) 100 mg/d (Zeffix, Glaxo Welcome, United Kingdom). After 18 to 24 mo, they all presented mutations conferring resistance to treatment. Owing to their similarities in LVD non-response, they were selected for inclusion in the study. To evaluate baseline variability, a sample taken at the time of the diagnosis (antiviral treatment naïve) was selected for each patient. HBV DNA was retested in all samples by TaqMan real-time polymerase chain reaction (PCR) (Roche) technology, and all presented values higher than 5 log10 IU/mL.
| Patient | Sex | Age (yr) | ALT (IU/mL) | HBV DNA (log10 IU/mL) | Genotype | HBeAg status |
| 1 | Male | 46 | 167 | 7.4 | A | -1 |
| 2 | Male | 39 | 95 | 7.8 | A | + |
| 3 | Male | 31 | 392 | 8.3 | D | - |
| 4 | Female | 55 | 117 | 7.5 | D | - |
Two sequential samples from patient 4 were additionally selected for UDPS analysis. HBV-DNA had been quantified using the branched-DNA (bDNA, limit of detection 5 logs IU/mL) technology available at that time, but the samples selected were retested with TaqMan technology for this study. At the time of the diagnosis (baseline sample), high HBV-DNA levels were detected by bDNA (retesting with TaqMan, 7 log10 IU/mL). However, HBV-DNA spontaneously dropped below the limit of detection of bDNA technology and the patient remained untreated, according to the guidelines at that time. After 36 mo (treatment-free sample), bDNA significantly increased (> 8 logs10 IU/mL on TaqMan retesting) and LVD was started. After an initial suboptimal response (HBV DNA decrease to 4 logs), viral breakthrough (7 logs10 HBV-DNA) was observed after 18 mo, and the rtL180M and rtM204V HBV polymerase variants were selected. Ultimately, adefovir was added to LVD.
All the samples included in this study had HBV viral loads higher than 6 logs IU/mL. HBV-DNA was extracted from serum by QIAamp microspin columns (QIAamp DNA Mini Kit, QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. To obtain optimal amplification of HBV DNA, the process was optimized with two PCRs. To minimize the error rate of the PCR process (false nucleotide substitutions), high fidelity polymerase (Pfu Ultra-II, Stratagene, La Jolla, United States) was used. At the time the study was designed, the maximal amplicon length that could be analyzed by the FLX platform was 250 nucleotides; PCR primers were selected for amplification of a specific 210-bp HBV fragment, which included main epitopic regions (Th50-69 and B74-84). The first PCR primers were sense (position 1662-1681); 5’-C/TATAAG AGGACTCTTGGACT-3’ and anti-sense primers (position 2912-2931); 5’-TGTTCCCAA/GGAATAA/TGGTGA-3’. The nested primers included the recognition site for UDPS, in italics. The sequence of the sense primer (position 1997-2016) was 5’-GCCTCCCTCGCGCCATCAGACCGCCTCAGCTCT C/TTAT CG-3’, and the anti-sense primer (position 2178-2206) was 5’-GCCTTGC CAGCCCGCTCAGCCACAA/GAGTTGCCTGAA/GCTT-3’. PCR products were isolated from 0.9% agarose gel and quantified using Quan-iT Picogreen sDNA reagent (Invitrogen). Before the sequencing reaction, each amplicon was pooled to obtain a concentration of 4 × 106 molecules of the HBV region. This working solution was enriched with the capture beads needed for sequencing. After optimal enrichment, clonal amplification in beads was performed in forward and reverse directions (emPCR kits II and III, 454 Life Sciences). UDPS was carried out in the Genome Sequencer FLX system (454 Life Sciences). The HBV region analyzed covered amino acids 40 to 95 of the core gene.
A total of 122 813 sequences was obtained. Reads were acquired with forward and reverse sequences and were aligned according to the primer sequence (designed by our group). Initial raw data filtering was performed as previously reported[16-18,20-22].
After applying the Poisson-based statistical filter validated in a study from our group[21], the empirical distribution of mismatch errors determined by UDPS analysis of an HBV DNA clone from the same region yielded an average of 0.006%; however in 8 positions, errors were higher than 0.02% but lower than 0.05%. Therefore, the sensitivity of UDPS to detect mutations was primarily limited by the highest mismatch error rate in the HBV DNA clone of 0.05%, which is similar to the value recently obtained in UDPS amplicons including an internal sequence as a control[20]. Thus, the measurements and biological conclusions in this study are based only on mutations present at a percentage above 0.05%.
Cloning of a more than full-length HBV genome[20] in pTriEx-mod vector was performed as described by Durantel et al[23]. The influence on HBV viral replication of the most conserved position observed in UDPS analysis, codon 76, was analyzed by site-directed mutagenesis (Agilent Technologies, Stratagene, United States) according to the manufacturer’s instructions. The wild-type clone had an L in codon 76, which was changed to V or P to test the effect of maintaining or deeply altering the physical-chemical properties of core codon 76. The introduction of mutations was confirmed by direct sequencing, as previously described[20].
Huh7 human hepatoma cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% calf serum. Transfection of plasmids was performed as previously described[20] using Fugene-HD (Roche, Germany). The supernatant was used to quantify HBsAg (Architect, Abbot), HBeAg (Vitros, Johnson and Johnson), and HBV DNA (CobasTaqman, Roche) production. The results were statistically analyzed with the Student t test. DNA was extracted from the supernatant (QiagenAMP DNA Mini Kit, Qiagen, Germany) following the manufacturer’s instructions and used to evaluate HBV-DNA production. As has been previously described[20], to confirm that HBV-DNA detected after transfection was the result of HBV replication and not due to contamination from the HBV genome in the pTriEx-mod vector of the transfection experiments, the supernatant extractions were used in 1/10, 1/102, 1/103, and 1/104 dilutions and PCR amplification of HBV-DNA and pTriEx-mod was performed.
To obtain the percentage of amino acid variability in each sample, the total number of amino acid substitutions was divided by the total number of amino acids analyzed. This value gave the theoretical variability for each position, and was used to estimate the expected variability for the regions studied (the theoretical variability was multiplied by the length of the epitope: 20 for Th50-69, 11 for B 74-84, and 25 for the remaining positions).
Fisher’s exact test was used to evaluate possible relationships between the most variable codons (variability ≥ 1%) and their positions in the epitopic region or other regions. The Wilcoxon signed-rank test was used to compare the evolution of the codons in the sequential analysis.
The amplicon analyzed was limited to codons 40 to 95, which include the main Th50-69 and B74-84 epitopes. A total of 122 814 sequences corresponding to 4 baseline samples were analyzed, and 108 403 of them were validated by bioinformatics and Poisson filtering. A total of 61 499 sequences were from genotype A samples and the remaining from genotype D. Variability was analyzed attending to the percentage of changes in all codons of the amplicon, and the results obtained for each position are shown in Table 2.
| Codon | PatientN sequencesMaster AA | Pt 133245% var | Pt 228254% var | Pt 327156% var | Pt 419748% var | Median baseline variability |
| 40 | E | 0.17 | 0.00 | 0.27 | 36.27 | 9.18 |
| 41 | A134/S2 | 0.04 | 7.54 | 1.58 | 0.03 | 2.30 |
| 42 | L | 0.20 | 0.08 | 0.14 | 0.09 | 0.13 |
| 43 | E | 0.06 | 0.07 | 0.08 | 0.12 | 0.08 |
| 44 | S | 0.03 | 0.01 | 0.01 | 0.05 | 0.03 |
| 45 | S1/P234 | 0.24 | 0.07 | 0.10 | 0.09 | 0.12 |
| 46 | E | 0.06 | 0.05 | 0.06 | 0.08 | 0.06 |
| 47 | H | 0.06 | 0.06 | 0.04 | 0.03 | 0.05 |
| 48 | C | 0.09 | 0.18 | 0.07 | 0.27 | 0.15 |
| 49 | S | 0.09 | 0.07 | 0.03 | 0.01 | 0.05 |
| 50 | P | 0.07 | 0.04 | 0.08 | 0.02 | 0.05 |
| 51 | H | 0.14 | 0.11 | 0.06 | 0.01 | 0.08 |
| 52 | H | 0.03 | 0.04 | 0.02 | 0.02 | 0.03 |
| 53 | T | 0.11 | 0.14 | 0.07 | 0.04 | 0.09 |
| 54 | A | 0.14 | 0.18 | 0.17 | 0.19 | 0.17 |
| 55 | L | 0.03 | 0.05 | 0.06 | 18.58 | 4.68 |
| 56 | R | 0.21 | 0.22 | 0.14 | 0.10 | 0.17 |
| 57 | Q | 0.05 | 0.05 | 0.03 | 0.00 | 0.03 |
| 58 | A | 0.15 | 0.18 | 0.09 | 0.04 | 0.12 |
| 59 | I134/V2 | 0.04 | 4.75 | 1.68 | 0.03 | 1.62 |
| 60 | L | 0.03 | 0.02 | 0.03 | 0.06 | 0.04 |
| 61 | C | 0.10 | 0.08 | 0.04 | 0.03 | 0.06 |
| 62 | W | 0.15 | 0.21 | 0.19 | 0.09 | 0.16 |
| 63 | G | 0.17 | 1.93 | 0.34 | 0.18 | 0.65 |
| 64 | D14/E23 | 0.69 | 0.50 | 0.41 | 29.96 | 7.89 |
| 65 | V1/L234 | 0.08 | 0.02 | 0.09 | 0.19 | 0.09 |
| 66 | T1/M234 | 0.12 | 0.21 | 0.11 | 0.11 | 0.14 |
| 67 | N1/T234 | 0.05 | 0.06 | 0.06 | 0.04 | 0.05 |
| 68 | L | 0.04 | 0.02 | 0.03 | 0.02 | 0.03 |
| 69 | A | 0.05 | 0.12 | 0.10 | 0.09 | 0.09 |
| 70 | T | 0.05 | 0.02 | 0.02 | 0.01 | 0.02 |
| 71 | W | 0.11 | 0.24 | 0.20 | 0.17 | 0.18 |
| 72 | V | 0.07 | 0.12 | 0.07 | 0.03 | 0.07 |
| 73 | G | 0.17 | 0.28 | 0.19 | 0.11 | 0.19 |
| 74 | N12/A3/V4 | 0.46 | 0.04 | 2.06 | 13.72 | 4.07 |
| 75 | N | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 |
| 76 | L | 0.00 | 0.01 | 0.02 | 0.02 | 0.01 |
| 77 | G134/Q2 | 0.14 | 0.30 | 1.94 | 3.03 | 1.35 |
| 78 | D | 0.03 | 0.08 | 0.03 | 0.08 | 0.05 |
| 79 | P | 0.16 | 0.10 | 0.07 | 0.03 | 0.09 |
| 80 | A123/T4 | 0.11 | 0.17 | 0.12 | 8.09 | 2.12 |
| 81 | S | 0.04 | 0.03 | 0.01 | 0.00 | 0.02 |
| 82 | R | 0.19 | 0.12 | 0.15 | 0.06 | 0.13 |
| 83 | D | 0.05 | 0.04 | 0.06 | 0.12 | 0.07 |
| 84 | Q1/L234 | 0.21 | 0.52 | 0.12 | 0.05 | 0.23 |
| 85 | V | 0.02 | 0.01 | 0.03 | 0.01 | 0.02 |
| 86 | V | 0.02 | 0.03 | 0.01 | 0.04 | 0.03 |
| 87 | N1/S234 | 0.06 | 0.03 | 1.93 | 0.02 | 0.51 |
| 88 | Y | 0.08 | 0.03 | 0.04 | 0.05 | 0.05 |
| 89 | V | 0.01 | 0.00 | 0.02 | 0.03 | 0.02 |
| 90 | N | 0.02 | 0.02 | 0.08 | 0.00 | 0.03 |
| 91 | T | 0.07 | 0.05 | 0.02 | 0.03 | 0.04 |
| 92 | N | 0.03 | 0.02 | 0.70 | 18.93 | 4.92 |
| 93 | M123/V4 | 0.02 | 0.05 | 0.15 | 18.89 | 4.78 |
| 94 | G | 0.04 | 0.12 | 0.15 | 0.09 | 0.10 |
| 95 | L | 0.03 | 0.02 | 0.15 | 0.14 | 0.09 |
In the two genotype A samples (patients 1 and 2), differences between the master sequences were found in ten codons (41, 45, 59, 64, 65, 66, 67, 77, 84 and 87, Table 2), seven of which were located in Th50-69 or B74-84. Of particular note, the master sequence of the motif delimited by codons 64 to 67, commonly defined by E64LMT67 and recognized by T-cells[13], differed in patient 1. The sequence found, D64VTN67, was completely different from the consensus sequence of genotypes A and D. The amino acid variability detected in patient 1 (average, 0.1%), which ranged from 0.69% to values under the cut-off (< 0.05%), was the lowest in all 4 samples. In this patient, the main epitopic regions contained 67.7% of the changes, a percentage 1.2 times higher than would be expected by the length of these regions, and the changes were equally distributed between the two epitopes. In contrast, patient 2 had higher variability (average, 0.35%), particularly in codons 41 (7.54%), 59 (4.75%) and 63 (1.93%). Only 53.1% of these changes were located in epitopic regions, a rate similar to the expected random percentage, but in the Th50-69 epitope the substitutions were 1.3 times higher than would be expected. Interestingly, two of the main substitutions detected in patient 2, S41 (A, 7.51%) and V59 (I, 4.21%), coincided with the master sequence of patient 1. The third main variant position was G63 (V, 1.62%).
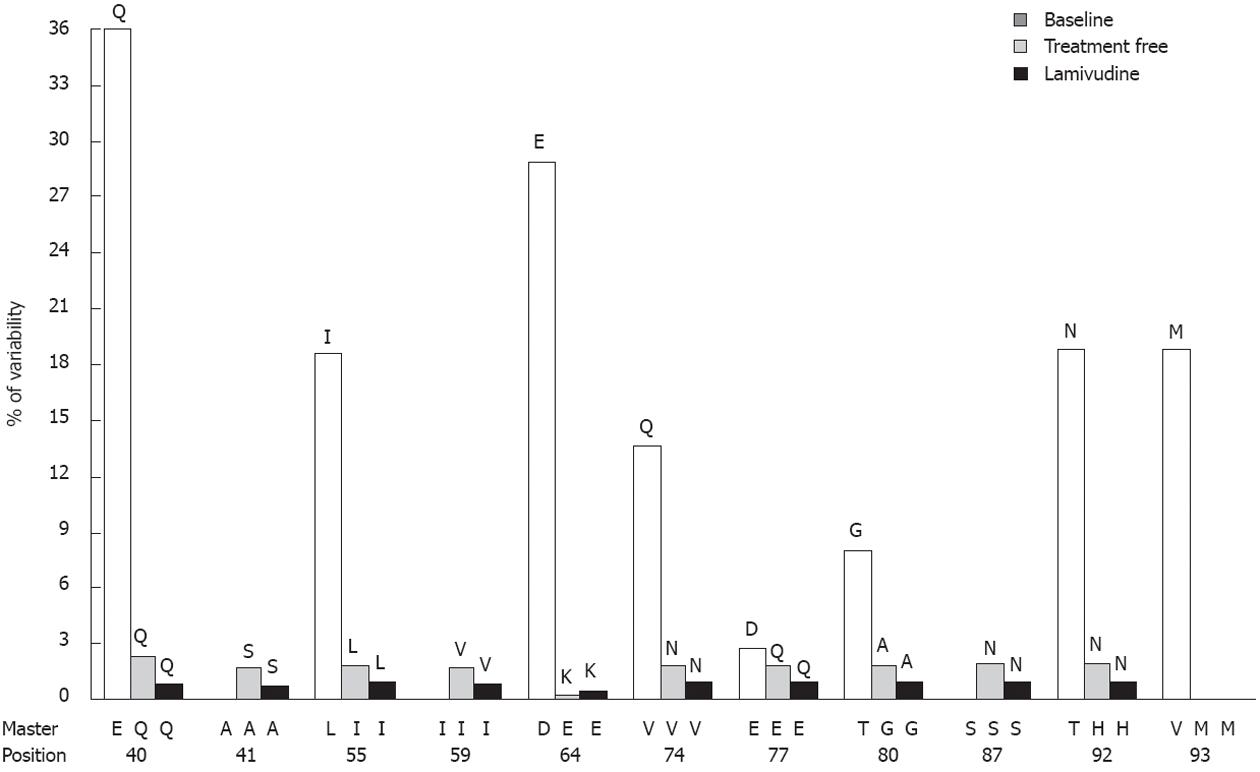
The two genotype D baseline samples (patients 3 and 4) had the same master sequence, except in codons 64, 74, 80 and 93, which were also the most highly variable in patient 4. In patient 3, five codons with more than 1% variability were detected: A41 (1.58%), I59 (1.68%), A74 (2.1%), E77 (1.94%) and S87 (1.93%) (3 of them in epitopic regions). The average amino acid variability was 0.26%, and 57.8% of changes were located in the main epitopic regions. Overall, this percentage was not higher than expected; however, changes in B74-84 were 1.6 times higher than the expected random percentage (31.6% vs 19.7%). In patient 4, variability was higher than 1% in 8 codons - Q40 (36.27%), L55 (18.58%), D64 (29.96%), V74 (13.72%), E77 (3.03%), T80 (8.08%), T92 (18.93%) and V93 (18.89%) - 5 of which were in epitopic regions. Linkage analysis showed that some of the main variants seen in this patient (S at codon 41, V at 59, N at 74, E at 77 and N at codon 87) were located in the same viral strain (1.5% of quasispecies). This observation seems to indicate possible selection by the effect of immune pressure on the core gene. Surprisingly, despite the high total amino acid variability detected in patient 4 (2.69%), only 49.8% of changes affected positions located in main epitopic regions, a value lower than was expected in both Th50-69 and B74-84.
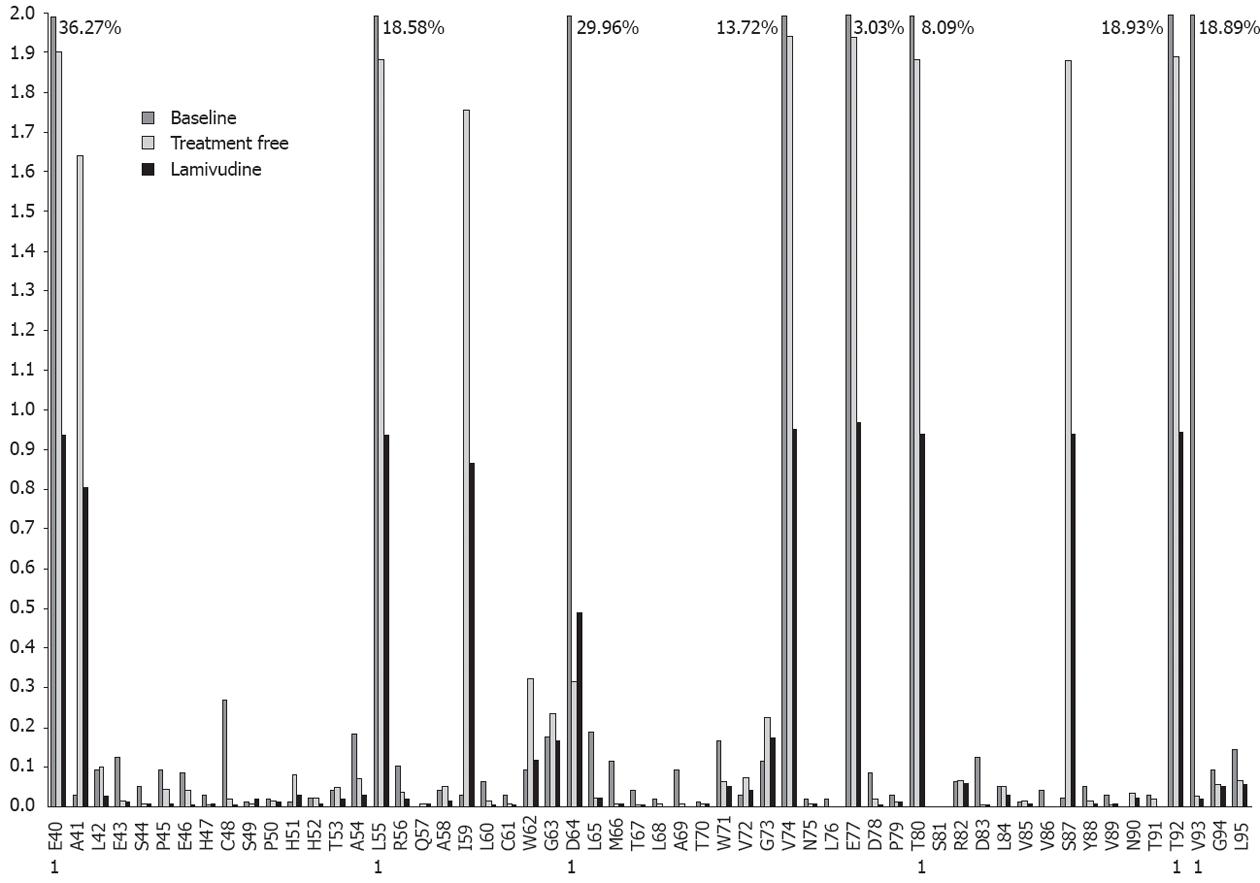
Median baseline frequencies were compared between the epitopic and other positions. Only genotype A samples showed high variability approaching significance (P = 0.07, sample 1 and P = 0.05, sample 2) in epitopic positions. Regarding the median baseline variability (Table 2), 6 of the highest values were located in positions within the main epitopic regions (codons 55, 59, 64, 74, 77 and 80), and 4 positions outside the main epitopes (codons 40, 41, 92 and 93) accumulated high percentages of changes. The variability in positions 40, 92 and 93 was due to changes in patient 4, whereas the variability of codon 41 (median 2.3% of changes) was due to changes in patients 2 and 3. Interestingly, positions 64 and 66, corresponding to the E64LMT67 motif of Th50-69, showed significant variability in all 4 samples (0.69%, 0.5%, 0.41% and 29.96% in position 64 and 0.12%, 0.21%, 0.11% and 0.11% in position 66).
Attending to the conserved positions, 12 codons showed variability lower than the system error rate (< 0.05%): positions 44, 52, 57, 68, 70, 75, 76, 78, 81, 85, 86 and 89. The most highly conserved was leucine in codon 76, with frequencies clearly below the system error rate (0.003-0.02%) and a median baseline error of 0.013% (Table 2). Based on this finding, codon 76 was analyzed by site-directed mutagenesis analysis.
Patient 4 was selected for sequential analysis, and 3 samples were processed (Figure 1): a baseline sample (also included in the Baseline Study), a sample following a treatment-free period of 36 mo, and a sample following 18 mo of LVD non-response. After application of the bioinformatic filter, 34 320 sequences from the treatment-free sample and 43 257 sequences from the LVD sample were obtained. The average amino acid variability of the baseline sample was higher than that of the treatment-free one (2.69% vs 0.34%, P = 0.001) and the average amino acid variability of the treatment-free sample was higher than that of the LVD treatment sample (0.34% vs 0.18%, P = 0.001). The main variants detected by sequential analysis are represented in Figure 2. Of note, 6 of these 11 variable positions (codons 55, 57, 64, 74, 77, 80) were located in main core epitopic regions.
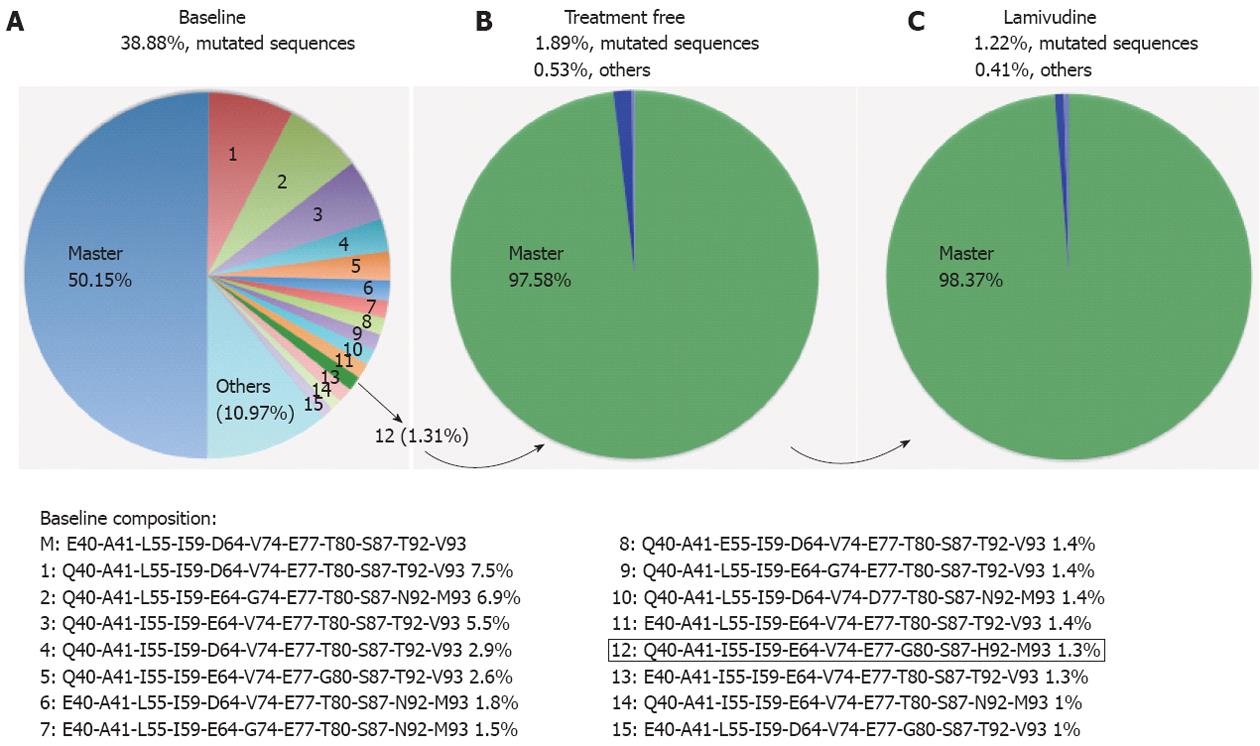
Linkage analysis was performed to determine whether the most frequent amino acid substitutions were simultaneously present in the same viral sequence (Figure 3A). At baseline, 50.15% of sequences in the positions studied were wild type, and 38.88% were mutated sequences in the most variable positions (15 different mutated variants, Figure 3A); 10.97% showed mutations in other positions. The mutated sequences in highly variable positions detected at baseline were found to be decreased in the treatment-free period (1.85%, Figure 3B) and after LVD breakthrough (1.24%, Figure 3B).
Attending to these variable positions, the most common strain at baseline (7.5%) had only one mutated codon (E40Q), followed by a strain (6.84%) with 5 mutated codons (E40Q, D64E, V74G, T92N and V93M) and another strain (5.45%) with 3 mutated codons (E40Q, L55I and D64E). Surprisingly, the baseline strain that had been selected as master after the treatment-free period and maintained during LVD was a low-frequency baseline mutant strain (1.31%) with the following substitutions: E40Q, L55I, D64E, T80G, T92H and V93M (variant 12, Figure 3A).
The time period with an absence of therapy (between baseline and treatment-free sampling) represented the complete time of HBV infection, and the HBV core quasispecies showed a tendency to decreased variability. This was reflected by a drop in the percentage of accumulated variability from baseline (38.88%) to the treatment-free sample (1.85%) and coincided with the change in the master sequence between the two samples, which could indicate a possible alternative immune escape mechanism. No significant differences were observed between samples from the treatment-free and LVD periods, which showed similar composition and percentages of mutated variant strains (Figure 3).
As is described above, leucine (L) at codon 76 was the most highly conserved position in all the samples analyzed (Table 2). Although there were other conserved positions (Q57, T70, D78 and S81), codon 76 focused our interest because it was a leucine (one amino acid coded by 6 different codons), because of its location in the core gene, and because it has never been described as essential. L76 was even more conserved than D78 (0.05%) or P79 (0.09%), both of which are reported to be involved in the core-HBsAg interaction[24]. Mutagenic studies of L76 were performed to evaluate a possible essential role of this amino acid. The experiments included a change to valine (V), whose hydrophobicity is similar to that of L, and a change to proline (P), whose physical-chemical properties differ from those of L.
After transfection, HBsAg, HBeAg and HBV DNA were quantified in cell culture supernatants. The presence of P in position 76 significantly decreased production of HBsAg, HBeAg and HBV-DNA in comparison with the wild type (L76) (both, P < 0.001). However, when V was in position 76, a reduction was observed in HBsAg levels (P < 0.001), but not in HBV DNA. Surprisingly, the V substitution resulted in a four-fold increase in HBeAg production in comparison with L76 (P < 0.001).
Several epitopic regions have been described in HBV core gene. Core region variability under antiviral treatment has been extensively studied, particularly in relation to IFN therapy[10,12]. However, differing patterns of amino acid substitutions under the effect of nucleotide/nucleoside analogues and during periods without treatment have been recently described[11]. Some studies have used the classical clonal method to analyze the evolution and composition of the entire HBV core gene[25-27]. However, in all these reports, only a small number of clones were processed and minor populations could not be studied. Thus, high-resolution clonal studies are needed to deeply analyze the composition of the HBV core gene quasispecies. To this end, the recently developed UDPS technology provides an opportunity to bypass the restrictions of the classical clonal method to determine the composition of viral quasispecies[16,17,19-21].
When this study was designed, UDPS technology based on the 454 Life Science Platform (GS-FLX, Roche Applied Science) only allowed analysis of around 200 nucleotide sequences. Because of this limitation, the present work was focused on analysis of the main immunodominant core motifs, Th50-69 and B74-84[6,7]. The high economic cost and complex computing analysis of UDPS strongly restrict the number of samples to be processed. For this reason, we were only able to study six selected samples: four samples to evaluate the baseline variability and two more samples to sequentially analyze one patient.
To achieve the main aim of UDPS analysis, a parallel analysis was needed to define the cut-off for analyzing the UDPS data. In our previous report of UDPS analysis of the HBV quasispecies[20], inclusion of an internal control sequence within the analyzed amplicon enabled establishment of a cut-off percentage to define the limit of viral variability, which was set at 0.03%. In the present study, a Poisson computational model recently validated by our group was applied by processing an HBV-DNA clone[21]; the percentage established to differentiate variability from UDPS error was set at 0.05%[21].
In the study of baseline HBV quasispecies, the most variable codons (median baseline variability ≥ 1%) were 40, 41, 55, 59, 64, 74, 77, 80, 92 and 93, all of which have been previously described in a study using clonal methodology to analyze acute exacerbations in HBeAg-negative patients[25]. Furthermore, some of these highly variable codons (codons 64, 74 and 77) were previously described as common changes in untreated chronic hepatitis B[11]. Genotype A sequences showed fewer codons with variability ≥ 1% and, consequently, lower overall median variability than genotype D. The statistical analysis of the median baseline frequencies showed that genotype A samples had higher variability in epitopic positions, approaching significance (P = 0.07, sample 1 and P = 0.05, sample 2). In contrast, in genotype D cases, no significant differences in variability were observed between the epitopic and other positions. These findings could be related to the high frequencies of mutations in codon 40, 87, 92 and 93 in genotype D samples (patients 3 and 4), which might be involved in the minor epitopic regions Th28-47 and Th82-101[6,11]. These differences in HBV core gene variability seem to indicate an influence of genotype on immune activation.
Patient 1 showed the lowest variability, but interestingly, the codon 64 to 67 master sequence was defined by the sequence D64VTN67, which is completely different from the well-characterized E64LMT67 motif. This motif is commonly recognized by T-cells, and the simultaneous E64D and T67N change has been reported to reduce T-cell proliferation in vitro[13]. Hence, we suggest that the changes observed in patient 1 might be the result of an alternative mechanism to escape from immune pressure, over the E64LMT67 motif. In fact, the finding that positions 64 and 66 of this motif showed significant variability in all 4 samples suggests that the motif could be a central target for immune pressure in HBV infection[11]. The low variability observed in sample 1 (0.1% of total amino acid variability) could reflect attainment of a kind of “steady state” in the viral quasispecies, resulting from strong selection of an escape variant with the mutated motif D64VTN67, similar to that seen in the longitudinal study of patient 4.
Evolution of the HBV quasispecies was evaluated in a single patient and involved a baseline sample and two additional samples, one taken after a period without treatment and one taken after LVD treatment. During the treatment-free period, a decrease in quasispecies variability was observed, with a reduction in the number of viral strains. This change may have been a consequence of host immune pressure[25]. One of the baseline viral strains present in a small percentage (variant 12, 1.31%, Figure 3) was strongly selected and became the master in the treatment-free and LVD samples, a fact suggesting that this variant might be an escape mutant, whose selection could be related to better fitness, regardless of its initial frequency.
Based on these results, we postulated that the HBV quasispecies achieved a kind of “steady state” after the treatment-free period that did not change with LVD treatment, because immune pressure could also be decreased during treatment. The significant differences in average variability in the two periods suggest that the equilibrium is dynamic. The structure of the HBV quasispecies in the three samples represents a complex reservoir of different minor variants, resulting from natural HBV evolution and likely affected by antiviral treatment[28].
Several codons in our region were found to be highly conserved (positions 57, 70, 76, 78 and 81). Position 76, located at the tip of the spike was of special interest, being next to the major immunodominant region and part of the B74-84 main epitope. Indeed, this position has never been described as essential, in contrast to the nearby positions 78 and 79[24]. The HBV core sequence is involved in the process of capsid conformation by interacting with the surface antigens in viral particle assembly. However, the interaction between HBsAg and HBcAg in virions is still unclear[29-31]. The core region analyzed in the present study is part of the assembly domain (amino acids 1-149); thus, the changes in this region might potentially modify the shell conformation.
Electrostatic interactions between core and HBsAg take place at the tip of the spike of the core antigen (codons 74-84)[32,33]. Cryo-electron microscopy studies have shown that codon 78 and 79 are within the contact area of core with envelope proteins[24]. This essential role might explain the high level of conservation of these positions (especially codon 78, 0.05%) observed in our UDPS analysis. However, surprisingly, the most conserved codon of the HBV core region in baseline and sequential samples was leucine at position 76.
To our knowledge, few studies[30,31] have analyzed the effect of single core amino acid mutations on HBV replication in vitro. Only one such study conducted by Ponsel et al[31] evaluated the leucine 76 position (among others) by inducing a change to alanine; no significant reduction in nucleocapsid or virion production was observed. In the present study, we induced changes in the hydrophobic characteristics of position 76 and determined their effect on HBsAg, HBeAg and HBV DNA production. In contrast to the results of Ponsel et al[31], we found a significant reduction in HBsAg production with both the V and P changes, suggesting possible involvement of L76 of HBcAg in the HBsAg interaction. A significant decrease in HBV replication in the presence of P76 was detected, leading us to speculate that the hydrophobic characteristics of position 76, conferred by the presence of L or V, are needed for HBV replication. The increase in HBeAg production in the presence of the V mutation and the absence of increasing HBV DNA were surprising, particularly because some authors have reported a correlation between HBeAg and HBV DNA levels[34]. Nonetheless, this correlation was not found by other authors[35] and currently remains controversial. We suggest that this unexpected HBeAg increase may indicate alternative pathways between HBeAg and HBV replication, as has been indicated previously[36]. Based on our in vitro results, it can be postulated that the amino acid changes induced in the core sequence are not as important as the structure adopted by the capsid and HBeAg.
Our study is mainly limited by the high cost of the UDPS process, which restricted the number of samples studied and created a risk that some results could be due to random chance. A larger number of samples, additional sequential studies, and duplicate UDPS experiments would have given more information about the HBV core sites involved in quasispecies evolution and potentially related to HBV chronic infection and to treatment non-response. In addition, at the time of the study, the available UDPS methodology only allowed analysis of sequences up to 250 bp. For this reason, we limited the study to the widely described main epitopic regions included in the HBV core, previously investigated by conventional methodologies[6,7,11,25].
In conclusion, this study validates application of UDPS to study the variability of the main core epitopes, substantiates the significant richness of the HBV baseline quasispecies, and suggests a relationship between core variability and HBV genotype. The highest variability was mainly detected in Th50-69 and B74-84, supporting their role as the main immune-stimulating core regions. The significant variability associated with well-characterized Th-cell motifs, such as E64LMT67, seems to indicate that the host immune system may be the main factor responsible for HBV core evolution. The relevant conservation of codon 76 may be related to possible interactions with the viral envelope. In the single longitudinally analyzed patient, a minor variant present in the baseline quasispecies was selected as the main variant in the absence of treatment and was maintained after lamivudine. These findings indicate the utility of UDPS to describe the dynamic behavior of the HBV quasispecies. More extended analyses with a larger number of samples must be performed to confirm the findings. The expected spread of this technology will probably allow a significant decrease in the cost, enabling processing a large number of samples. The UDPS application for diagnostic and routine analysis might serve for the quantitative estimation of the viral quasispecies, for defining mutant variants or for establishing the quasispecies complexity. All these parameters would be useful as prognostic factors for disease outcome or therapy efficacy.
The authors thank Celine Cavallo for English language support and helpful editing suggestions.
Hepatitis B virus (HBV) infection is a global health problem, with more than 300 million HBV carriers. Viral distribution in individual infection is defined as a quasispecies, which means that variability can be accumulated along the HBV genome. The core gene is the least overlapped gene of the HBV genome and core antigen is the most immunogenic of all the viral peptides. Therefore, the region encoding the main core epitopes is useful to study the effect of the host immune response and to analyze the variability of HBV quasispecies composition.
The distribution of viral infection as a quasispecies (in HBV, but also hepatitis C virus and human immunodeficiency virus) is an important advantage for the virus. It enables fast, easy establishment of infection and enables adaptation against changes in the viral environment. The study of viral quasispecies has been limited by the available methods, with which analysis of a significant number of clonal sequences was extremely difficult. The possibility of obtaining thousands of sequences in a single sample, provided by next-generation sequencing methods such as ultra-deep-pyrosequencing (UDPS), will allow more in-depth study of the viral quasispecies. In the present study, the authors applied the UDPS to analyze HBV quasispecies variability and adaptability of the virus in single patient.
To date, few studies have used UDPS to study the HBV quasispecies, likely because of the current high cost of the technique for this purpose. However, the results presented in these recently published studies have shown clear advantages of UDPS for viral quasispecies analysis. Most of these studies have been mainly focused on the polymerase gene (treatment resistance variants). The present study is the first work analyzing the core gene by this method, to evaluate the effect of the host immune response and the variability of this little overlapped region of the HBV genome.
This study illustrates the value of deep quantitative analysis of the HBV quasispecies composition to investigate its clinical relevancy. The results provide an indication of the role of HBV core gene quasispecies structure in the natural host immune response and have prompted us to continue investigating HBV variability by UDPS, particularly the precore and core regions because of their relationship with the immune response.
Viral quasispecies distribution in an infected patient refers to a group of viruses that are different, but highly related. This distribution results in competition between viruses from the same infection, but also confers plasticity and adaptability to environmental changes.
The authors analyzed the quasispecies of HBV core epitopic regions by UDPS. They found that positions with highest variability rates, mainly clustered in the main core epitopes, showed some relationship with HBV genotype, and were particularly associated with the T-helper motif. The authors suggest that immune system pressure is the main cause of HBV core evolution.
Peer reviewers: Eric R Kallwitz, MD, Assistant Professor, Department of Medicine, University of Illinois, 840 S Wood Street, 7th Floor, MC 787, Chicago, IL 60612, United States; Andi Utama, PhD, Molecular Epidemiology Division, Mochtar Riady Institute For Nanotechnology, Jl. Boulevard Jend. Sudirman 1688, Lippo Karawaci-Tangerang, Banten 15810, Indonesia
S- Editor Gou SX L- Editor O’Neill M E- Editor Li JY
| 1. | Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 317] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 2. | Ehata T, Omata M, Yokosuka O, Hosoda K, Ohto M. Variations in codons 84-101 in the core nucleotide sequence correlate with hepatocellular injury in chronic hepatitis B virus infection. J Clin Invest. 1992;89:332-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 121] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Preikschat P, Günther S, Reinhold S, Will H, Budde K, Neumayer HH, Krüger DH, Meisel H. Complex HBV populations with mutations in core promoter, C gene, and pre-S region are associated with development of cirrhosis in long-term renal transplant recipients. Hepatology. 2002;35:466-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Sung FY, Jung CM, Wu CF, Lin CL, Liu CJ, Liaw YF, Tsai KS, Yu MW. Hepatitis B virus core variants modify natural course of viral infection and hepatocellular carcinoma progression. Gastroenterology. 2009;137:1687-1697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Vanlandschoot P, Cao T, Leroux-Roels G. The nucleocapsid of the hepatitis B virus: a remarkable immunogenic structure. Antiviral Res. 2003;60:67-74. [PubMed] |
| 6. | Ferrari C, Bertoletti A, Penna A, Cavalli A, Valli A, Missale G, Pilli M, Fowler P, Giuberti T, Chisari FV. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J Clin Invest. 1991;88:214-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 171] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Carman WF, Boner W, Fattovich G, Colman K, Dornan ES, Thursz M, Hadziyannis S. Hepatitis B virus core protein mutations are concentrated in B cell epitopes in progressive disease and in T helper cell epitopes during clinical remission. J Infect Dis. 1997;175:1093-1100. [PubMed] |
| 8. | Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398-1401. [PubMed] |
| 9. | Bozkaya H, Ayola B, Lok AS. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology. 1996;24:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Radecke K, Protzer U, Trippler M, Meyer Zum Büschenfelde KH, Gerken G. Selection of hepatitis B virus variants with aminoacid substitutions inside the core antigen during interferon-alpha therapy. J Med Virol. 2000;62:479-486. [PubMed] |
| 11. | Homs M, Jardi R, Buti M, Schaper M, Tabernero D, Fernandez-Fernandez P, Quer J, Esteban R, Rodriguez-Frias F. HBV core region variability: effect of antiviral treatments on main epitopic regions. Antivir Ther. 2011;16:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Naoumov NV, Thomas MG, Mason AL, Chokshi S, Bodicky CJ, Farzaneh F, Williams R, Perrillo RP. Genomic variations in the hepatitis B core gene: a possible factor influencing response to interferon alfa treatment. Gastroenterology. 1995;108:505-514. [PubMed] |
| 13. | Torre F, Cramp M, Owsianka A, Dornan E, Marsden H, Carman W, Williams R, Naoumov NV. Direct evidence that naturally occurring mutations within hepatitis B core epitope alter CD4+ T-cell reactivity. J Med Virol. 2004;72:370-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Alexopoulou A, Karayiannis P, Hadziyannis SJ, Aiba N, Thomas HC. Emergence and selection of HBV variants in an anti-HBe positive patient persistently infected with quasi-species. J Hepatol. 1997;26:748-753. [PubMed] |
| 15. | Jiang Y, Li W, Yu L, Liu J, Xin G, Yan H, Sun P, Zhang H, Xu D, Niu J. Enhancing the antihepatitis B virus immune response by adefovir dipivoxil and entecavir therapies. Cell Mol Immunol. 2011;8:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, Holmes SP, Babrzadeh F, Gharizadeh B, Hanczaruk B. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J Infect Dis. 2009;199:1275-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 17. | Solmone M, Vincenti D, Prosperi MC, Bruselles A, Ippolito G, Capobianchi MR. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J Virol. 2009;83:1718-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 130] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 18. | Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 318] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 19. | Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195-1201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 344] [Cited by in RCA: 321] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 20. | Homs M, Buti M, Quer J, Jardí R, Schaper M, Tabernero D, Ortega I, Sanchez A, Esteban R, Rodriguez-Frias F. Ultra-deep pyrosequencing analysis of the hepatitis B virus preCore region and main catalytic motif of the viral polymerase in the same viral genome. Nucleic Acids Res. 2011;39:8457-8471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Rodriguez-Fras F, Tabernero D, Quer J, Esteban J, Ortega I, Domingo E, Cubero M, Cams S, Ferrer-Costa C, Snchez A. Ultra-Deep Pyrosequencing Detects Conserved Genomic Sites and Quantifies Linkage of Drug-Resistant Amino Acid Changes in the Hepatitis B Virus Genome. PloS One. 2012;7:e37874. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 914] [Cited by in RCA: 857] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 23. | Durantel D, Carrouée-Durantel S, Werle-Lapostolle B, Brunelle MN, Pichoud C, Trépo C, Zoulim F. A new strategy for studying in vitro the drug susceptibility of clinical isolates of human hepatitis B virus. Hepatology. 2004;40:855-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Seitz S, Urban S, Antoni C, Böttcher B. Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions. EMBO J. 2007;26:4160-4167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Alexopoulou A, Baltayiannis G, Eroglu C, Nastos T, Dourakis SP, Archimandritis AJ, Karayiannis P. Core mutations in patients with acute episodes of chronic HBV infection are associated with the emergence of new immune recognition sites and the development of high IgM anti-HBc index values. J Med Virol. 2009;81:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Osiowy C, Kaita K, Solar K, Mendoza K. Molecular characterization of hepatitis B virus and a 9-year clinical profile in a patient infected with genotype I. J Med Virol. 2010;82:942-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Tang YZ, Liu L, Pan MM, Wang YM, Deng GH. Evolutionary pattern of full hepatitis B virus genome during sequential nucleos(t)ide analog therapy. Antiviral Res. 2011;90:116-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 28. | Domingo E, Gomez J. Quasispecies and its impact on viral hepatitis. Virus Res. 2007;127:131-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 29. | Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771-780. [PubMed] |
| 30. | Pairan A, Bruss V. Functional surfaces of the hepatitis B virus capsid. J Virol. 2009;83:11616-11623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77:416-422. [PubMed] |
| 32. | Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269-4274. [PubMed] |
| 33. | Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 293] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 34. | Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, Iinuma K, Mushahwar IK. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J Virol Methods. 2004;115:217-222. [PubMed] |
| 35. | Wiegand J, Wedemeyer H, Finger A, Heidrich B, Rosenau J, Michel G, Bock CT, Manns MP, Tillmann HL. A decline in hepatitis B virus surface antigen (hbsag) predicts clearance, but does not correlate with quantitative hbeag or HBV DNA levels. Antivir Ther. 2008;13:547-554. [PubMed] |
| 36. | Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 339] [Article Influence: 22.6] [Reference Citation Analysis (0)] |