Published online Mar 28, 2012. doi: 10.3748/wjg.v18.i12.1397
Revised: September 30, 2011
Accepted: January 7, 2012
Published online: March 28, 2012
AIM: To investigate whether Notch signaling is involved in liver fibrosis by regulating the activation of hepatic stellate cells (HSCs).
METHODS: Immunohistochemistry was used to detect the expression of Notch3 in fibrotic liver tissues of patients with chronic active hepatitis. The expression of Notch3 in HSC-T6 cells treated or not with transforming growth factor (TGF)-β1 was analyzed by immunofluorescence staining. The expression of Notch3 and myofibroblastic marker α-smooth muscle actin (α-SMA) and collagen I in HSC-T6 cells transfected with pcDNA3.1-N3ICD or control vector were detected by Western blotting and immunofluorescence staining. Moreover, effects of Notch3 knockdown in HSC-T6 by Notch3 siRNA were investigated by Western blotting and immunofluorescence staining.
RESULTS: The expression of Notch3 was significantly up-regulated in fibrotic liver tissues of patients with chronic active hepatitis, but not detected in normal liver tissues. Active Notch signaling was found in HSC-T6 cells. TGF-β1 treatment led to up-regulation of Notch3 expression in HSC-T6 cells, and over-expression of Notch3 increased the expression of α-SMA and collagen I in HSC-T6 without TGF-β1 treatment. Interestingly, transient knockdown of Notch3 decreased the expression of myofibroblastic marker and antagonized TGF-β1-induced expression of α-SMA and collagen I in HSC-T6.
CONCLUSION: Notch3 may regulate the activation of HSCs, and the selective interruption of Notch3 may provide an anti-fibrotic strategy in hepatic fibrosis.
- Citation: Chen YX, Weng ZH, Zhang SL. Notch3 regulates the activation of hepatic stellate cells. World J Gastroenterol 2012; 18(12): 1397-1403
- URL: https://www.wjgnet.com/1007-9327/full/v18/i12/1397.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i12.1397
Hepatic fibrosis is a reversible wound-healing response characterized by the accumulation of extracellular matrix (ECM) to liver injury[1]. In the process of hepatic fibrosis, activated hepatic stellate cells (HSCs) synthesize a large amount of ECM and then change into myofibroblasts[2], which is characterized by the expression of α-smooth muscle actin (α-SMA) and ECM, particularly collagen I. Myofibroblast is one of the key cellular components involved in liver fibrosis, therefore, the majority of anti-fibrotic therapies are designed to inhibit the activation, proliferation, or synthetic products of HSCs[3].
Notch signaling is an ancient cell signaling that regulates cell fate specification, stem cell maintenance, and initiation of differentiation in embryonic and postnatal tissues[4,5]. More recently, some researches reported that Notch signaling was implicated in human fibrosis diseases, such as pulmonary, renal and peritoneal fibrosis[6-8]. Several researches suggested that the Jagged/Notch pathway may selectively mediate fibrogenic properties of transforming growth factor-β1 (TGF-β1) which was essential to promote the production and deposition of ECM[9-11].
Notch receptors (Notch1, Notch2 and Notch4) were present at the mRNA level in freshly isolated HSCs, and the synthesis of Notch1 decreased during culture and development of HSCs into myofibroblast-like cells[12]. However, the expression of Notch3 in phenotype activated HSCs remains unknown. Ono et al[13] reported that Notch3 was required for TGF-β1-induced myofibroblastic differentiation of myoblasts. Based on the studies above, the present study was undertaken to investigate whether Notch3 was expressed in fibrotic liver tissues of patients with chronic active hepatitis and in activated HSCs, and sequentially contributed to liver fibrosis by regulating the activation of HSCs.
Liver tissue samples were obtained by biopsy from 11 patients with chronic active hepatitis (5 women and 6 men; median age 43 years, range 31-55 years). Control liver biopsy specimens were obtained from healthy volunteers (n = 6). All patients and controls signed consent forms approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology. Tissue samples were fixed in 10% formalin and paraffin-embedded for immunohistochemical analysis.
Rabbit polyclonal to α-SMA and Notch3 were obtained from Abcam (Cambridge, United States). Rabbit polyclonal anti-collagen I antibody was purchased from Bioss Corporation (Beijing, China). Horseradish peroxidase (HRP)-conjugated anti-rabbit IgG was obtained from Santa Cruz Biotechnology (Santa Cruz, United States). Recombinant human transforming growth factor (TGF)-β1 was purchased from PeproTech EC Ltd (London, United Kingdom). Lipofectamine™ 2000 transfection reagent was obtained from Invitrogen (Carlsbad, United States).
HSC-T6 cells, an immortalized rat HSC line, purchased from Cancer Institute and Hospital, Chinese Academy of Medical Sciences (China), were cultured in Dulbecco’s-modified Eagle’s medium (DMEM; Gibco, United States) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% fetal bovine serum (FBS) (Gibco, United States). TGF-β1 (2 ng/mL) was incubated in growth medium for 24 h.
Liver tissue sections were incubated with 3% H2O2 followed by serum blocking with 10% goat serum in 5% bovine serum albumin (BSA). The Notch3 was detected by staining with polyclonal rabbit anti-Notch3 (1:250) overnight at 4 °C. Irrelevant isotype antibodies (Santa Cruz Biotechnology, United States) at the same concentration were used as control. The staining was carried out using SABC kit (Boster, China).
Cells were fixed in phosphate buffered saline (PBS) containing 4% paraformaldehyde at room temperature for 30 min and were penetrated in blocking solution (Amresco, United States) containing 0.3% Triton X-100, and incubated overnight with either anti-Notch3 (1:250), anti-α-SMA (1:100) or anti-collagen I (1:100) antibody in 1% BSA solution. Then, cells were incubated with secondary antibodies for 1 h at 37 °C. After incubation, cells were stained with nuclear stain marker 4’,6-diamidino-2-phenylindole. The reaction was examined under confocal microscope (Nikon, Japan).
HSC-T6 was seeded into 6-well plates at a density of 1 × 105 cells 12 h before transfection. siRNA was mixed with 5 μL lipofectamine 2000 in 250 μL Opti-MEM I medium for 20 min. The transfection mixture was then added to each well with 1.5 mL FBS free DMEM at a concentration of 100 nmol/L. After 6-h incubation, liquid mixture containing siRNA was disposed. Two mL DMEM containing 10% FBS was added and incubated for another 72 h. The following siRNA sequences were used: Notch3 siRNA: 5’ ACAAGAUCAAUACAGGAGCTT 3’; the control siRNA sequence: 5’ UUGUAC UACACAAAAGUACUG 3’. These siRNA were synthesized by Shanghai Genepharma Co. Ltd. (Shanghai, China).
HSC-T6 was transfected with Notch3 intracellular domain (Notch3-ICD) cDNA cloned into pcDNA3.1 (pc-DNA3.1-N3ICD) vector, which was a gift from Dr. Tao Wan. The control (pcDNA3.1-empty) vector was purchased from Shanghai Genepharma Co. Ltd. (Shanghai, China). All transfections were performed using Lipofectamine 2000 following the manufacturer’s instructions.
Total RNA from each sample was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. Real-time polymerase chain reaction (RT-PCR) was performed using a StepOne/StepOne-Plus (ABI) and the TaqMan PCR Reagent (Genepharma). Primer sequences are summarized in Table 1. Comparative threshold (Ct) method was used for calculating the relative amount of mRNA of treated sample compared with control samples.
| Notch3 | |
| Forward | 5’-CCTGCCTGCCTCTATGACAAC-3’ |
| Reverse | 5’-ACACTCCTCGGTGTTACAGCC-3’ |
| Probe | 5’-ACTGCTACTCTGGTGGCCGCGAC-3’ |
| Jagged1 | |
| Forward | 5’-GTGGAAGAGGATGATATGGATAAGC-3’ |
| Reverse | 5’-CTCCTCTCTGTCTACCAGCGTGTAC-3’ |
| Probe | 5’-CCAGCAGAAAGTCCGGTTTGCCA-3’ |
| Hes1 | |
| Forward | 5’-TGCTACCCCAGCCAGTGTC-3’ |
| Reverse | 5’-GCTTTGATGACTTTCTGTGCTCA-3’ |
| Probe | 5’-CTGTCTTTGGTTTGTCCGGTGTCGT-3’ |
| GAPDH | |
| Forward | 5’-GATGACATCAAGAAGGTGGTGAAG-3’ |
| Reverse | 5’-ACCCTGTTGCTGTAGCCATATTC-3’ |
| Probe | 5’-ACTTCAACAGCAACTCCCACTCTTCCACC-3’ |
Cells were washed with PBS and lysed. The extracts were cleared by centrifugation at 12 000 ×g for 15 min. After blocking with 5% non-fat milk in PBS containing 0.1% Tween 20 for 1 h at room temperature, membranes were incubated with either anti-Notch3 (1:1000), anti-α-SMA (1:300) or anti-collagen I (1:200) antibody in tris-buffered saline (TBS) containing 0.05% Tween 20 at 4 °C overnight. Then membranes were incubated with HRP-conjugated secondary anti-rabbit IgG (1:2000) antibody in TBS and Tween 20 for 1 h at room temperature, and visualized by chemiluminescence using an electrochemiluminescence immunoblotting kit (Cell Signaling Technology) with a digital luminescent image analyzer BioSpectrum600 (UVP, United States). Band intensity was assessed using Gel-Pro analyzer.
All experiments were repeated three times, and data recorded as mean ± SD and analyzed by Student’s t test using SPSS12.0 software. P < 0.05 was considered statistically significant.
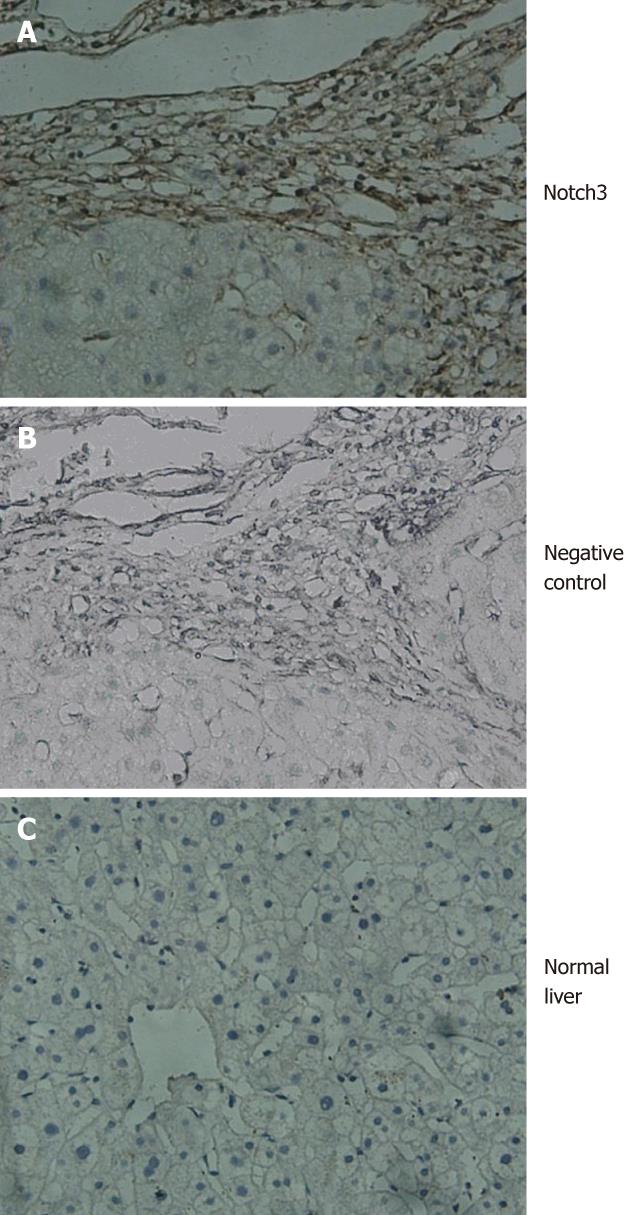
All patients were positive for the Notch3 in fibrotic liver tissues (Figure 1A). In contrast, Notch3 was not detected by immunohistochemistry in normal liver tissues (Figure 1C).
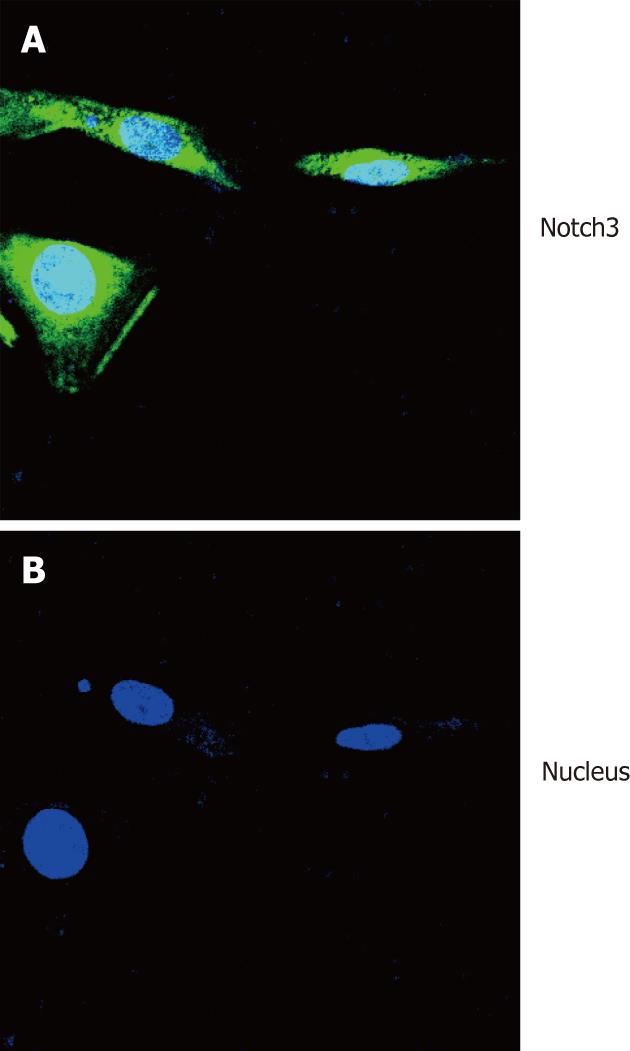
To detect expression of Notch3 in HSCs, immunofluorescence staining analysis was performed to examine the expression of Notch3 in HSC-T6 cells. The result showed that Notch3 protein was localized in the cytoplasm and nucleus of HSC-T6 cells (Figure 2).
Up-regulation of Notch3 expression by transforming growth factor-β1 in hepatic stellate cell-T6 cells
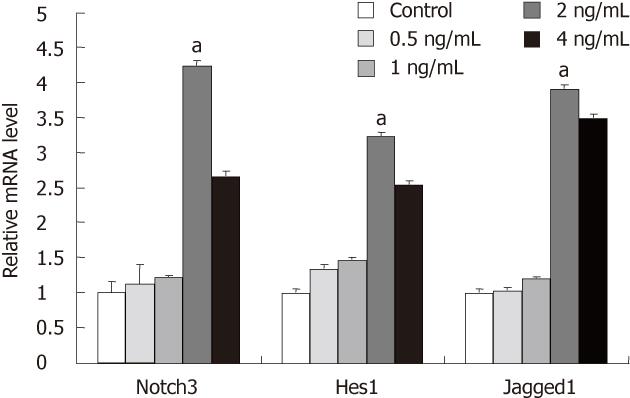
We investigated the effect of TGF-β1 on Notch signaling in HSC-T6 treated with TGF-β1 (0.5, 1, 2 and 4 ng/mL) for 24 h. RT-PCR analysis showed that the expression of Notch signaling components including Notch3, Jagged1 and Hes-1 were obviously increased in HSC-T6 treated with 2 ng/mL TGF-β1 as compared with the control group without TGF-β1 treatment (P < 0.05, Figure 3).
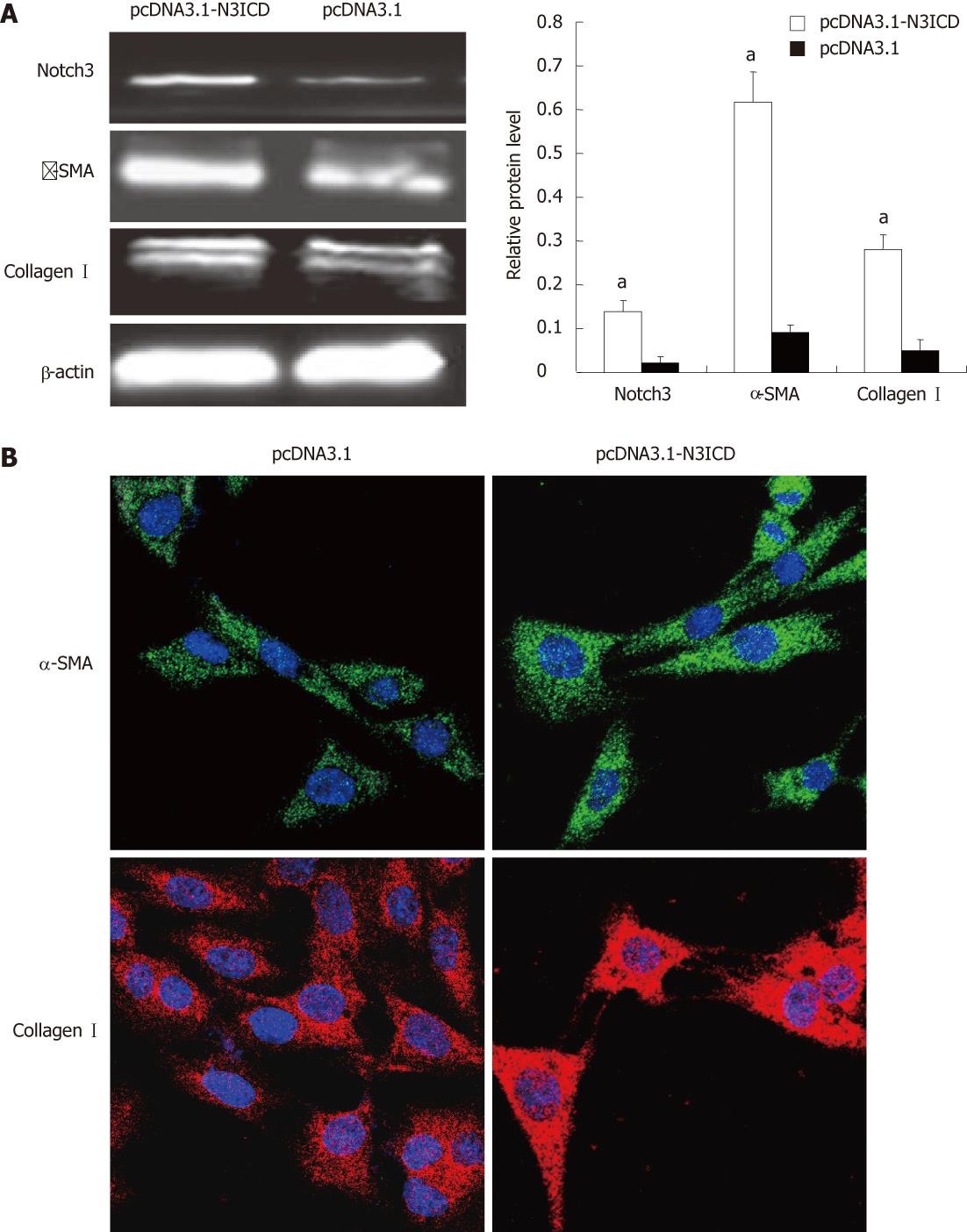
To investigate the effect of Notch3 in activation of HSCs, we examined if overexpresion of Notch3 in HSC-T6 would enhance the activation. The results showed that the increased expression of Notch3 in pcDNA3.1-N3ICD introduced HSC-T6 cells as compared with cells transfected with pcDNA3.1-empty vector (P < 0.05, Figure 4A). Western blotting and immunofluorescence staining analyses demonstrated that over-expression of Notch3 led to increased expression of α-SMA and collagen Icompared with control group (P < 0.05, Figure 4A and B).
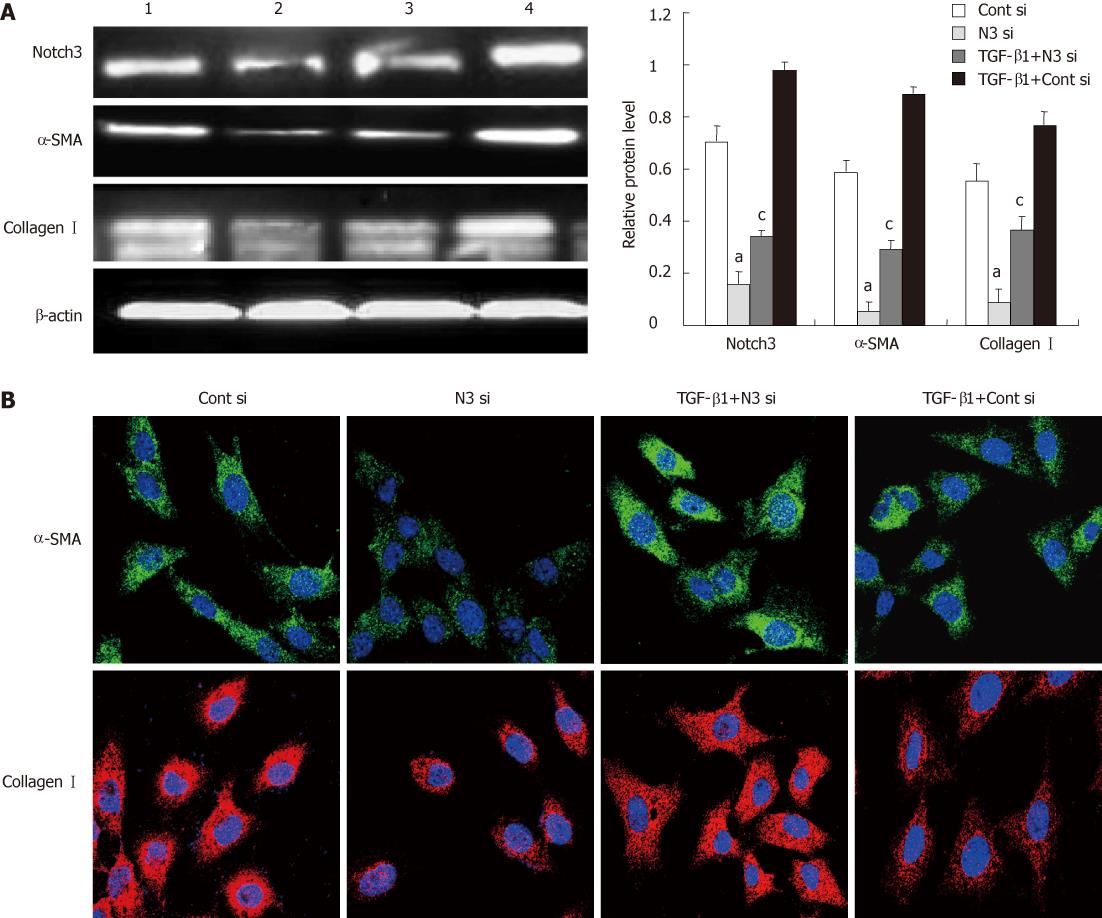
To further confirm the role of Notch3 in regulating activation of HSCs, siRNA was employed to specifically knockdown Notch3. Western blotting analysis showed that siRNAs targeting Notch3 reduced Notch3 protein levels by approximately 80%. Seventy and two hour after transfection in HSC-T6 (P < 0.05, Figure 5A). We also observed that knockdown of Notch3 in HSC-T6 down-regulated the expression of α-SMA and collagen I detected by Western blotting and immunofluorescence staining 72 h after siRNA transfection (P < 0.05, Figure 5A and B).
To investigate the relationship between Notch3 and TGF-β1, TGF-β1 (2 ng/mL) was added into HSC-T6 24 h before transfection with siRNAs targeting Notch3 or control siRNAs. Western blotting and immunofluorescence staining analyses demonstrated that knockdown of Notch3 antagonized TGF-β1-induced expression of α-SMA and collagen I in HSC-T6 (P < 0.05, Figure 5A and B).
Liver fibrosis is the result of the wound-healing response to repeated injury in liver. It is well known that HSCs activation plays an important role in fibrosis because these cells become the primary source of extracellular matrix in liver upon injury. TGF-β1 is known to promote fibrogenesis in vivo and in vitro, however, development of anti-fibrotic strategies targeting the TGF-β axis is problematic owing to the pleiotropic nature of TGF-β action[1].
Notch signaling is an evolutionarily conserved local cell-signaling that functions in the determination of cellular identity during developmental stages[14]. Four Notch proteins (Notch1, Notch2, Notch3 and Notch4) have been identified in vertebrates, while membrane-bound proteins (Delta and Jagged) have been recognized as Notch ligands[15]. Activation of the pathway usually occurs via expression of the ligand in signal-giving cells. Upon interaction with the ligand, Notch undergoes a series of proteolytic cleavages in the signal-receiving cells. Finally, the cytoplasmic domain, referred to as the Notch intracellular domain (NICD), translocates to the nucleus, where it binds with the transcription factor CSL [CBF-1/RBP-Jk, Su (H) and Lag-1] and co-activator Mastermind-like to trigger downstream target genes expression, such as HES1 and HEY which act as transcription factors[16-20].
Notch signaling is essential for the regulation of cell differentiation, and its aberrant activation was implicated in human fibrosis diseases. Notch1 signaling in response to inflammatory zone 1 may play a significant role in myofibroblast differentiation during lung fibrosis[6]. Active Notch pathway in tubular epithelial cells was demonstrated as a critical regulator of tubulointerstitial fibrosis[7]. It was reported that Notch signaling was highly activated in rats with fibrotic peritoneum induced by peritoneal dialysis fluid , as indicated by increased expression of Jagged1, Notch1, and HES1. Blocking Notch signaling activation by intraperitoneal injection of a γ-secretase inhibitor significantly attenuated peritoneal fibrosis as indicated by the decreased expression of α-smooth muscle actin and collagen I[8].
In this study, the Notch3 was not detected in normal liver tissues. In contrast, intense staining of the Notch3 was observed in fibrotic liver tissues of patients with chronic active hepatitis, which suggested that Notch3 was involved in liver fibrosis. Furthermore, several lines of evidence clearly confirmed that Notch3 contributed to liver fibrosis by regulating activation of HSCs. First, the expression of active Notch3 was found in the nucleus of HSC-T6. Second, the expression level of Notch signaling components was elevated including Notch3, Jagged1 and Hes1 in HSC-T6 after TGF-β1 treatment. More importantly, we also found that specific knockdown of Notch3 by siRNA antagonized TGF-β1-induced expression of myofibroblastic marker α-SMA and collagen I in HSC-T6, and on the contrary, over-expression of Notch3 increased the expression of myofibroblastic marker in HSC-T6.
Inhibition of the γ-secretase complex required for release of the active NICD is the most common method for targeting Notch signaling[21,22]. Two different inhibitors are being evaluated in clinical trials for the treatment of resistant T cell acute leukemia and advanced breast cancer (http://www.clinicaltrials.gov). However, targeting of γ-secretase would result in several adverse events when Notch receptors from Notch1 to Notch4 are affected[23,24]. In this study, we found that the selective interruption of Notch3 by siRNA decreased the expression of myofibroblastic marker in HSC-T6 cells, which provided a potential novel therapeutic target for liver fibrosis.
However, further studies are required to elucidate how do other members of Notch family, such as Notch1, Notch2 and Notch4, play a part in activated HSCs as well as the mechanism underlying the Notch signaling in liver fibrosis. In addition, the contribution of Notch and TGF-β1 signaling to the liver fibrosis should also be further investigated.
In conclusion, we demonstrated for the first time that Notch3 plays a role in regulating the activation of HSCs (HSC-T6). Therefore, the selective interruption of Notch3 may have a potential anti-fibrogenic effect in liver fibrosis.
It is well known that hepatic stellate cell (HSC) activation plays an important role in fibrosis because these cells are the primary source of extracellular matrix in liver upon injury. Notch signaling regulates many aspects of morphogenesis through diverse effects on differentiation, proliferation, and cell survival. Recently, some researches reported that Notch signaling was implicated in human fibrosis diseases, such as pulmonary, renal and peritoneal fibrosis.
This study was undertaken to investigate whether Notch signaling is activated in HSCs and sequentially contributed to liver fibrosis by regulating the activation of HSCs.
This is the first study to characterize the role of Notch signaling in liver fibrosis. This finding indicated that Notch3 may contribute to liver fibrosis by regulating the activation of HSCs.
This study showed that the selective interruption of Notch3 by siRNA decreased the expression of myofibroblastic marker in hepatic stellate cell-T6 cells, which provided a potential novel therapeutic target for liver fibrosis.
This is a very interesting study aimed at investigating the role of the Notch signaling pathway in liver fibrosis. The text is generally well written, with structured abstract and organized sections. The manuscript has scientific value since it includes original information about a relevant topic.
Peer reviewers: Eddie Wisse, Professor emeritus of Laboratory for Cell Biology and Histology (VUB), Hon. Professor of Internal Medicine, University of Maastricht, The Netherlands, and Hon. Professor of Australian Key Center for Microscopy and Microanalysis, University of Sydney, Irisweg 16, Keerbergen 3140, Belgium; Roberto J Carvalho-Filho, MD, PhD, Hepatitis Section, Division of Gastroenterology, Federal University of Sao Paulo, Rua Botucatu, 740, 2.o andar, Vila Clementino, State of Sao Paulo 04023-060, Brazil
S- Editor Shi ZF L- Editor Ma JY E- Editor Xiong L
| 1. | Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1377] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 2. | Wells RG. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J Clin Gastroenterol. 2005;39:S158-S161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 176] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21:437-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 397] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 4. | Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 264] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 5. | Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4480] [Cited by in RCA: 4552] [Article Influence: 175.1] [Reference Citation Analysis (0)] |
| 6. | Liu T, Hu B, Choi YY, Chung M, Ullenbruch M, Yu H, Lowe JB, Phan SH. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. Am J Pathol. 2009;174:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 7. | Bielesz B, Sirin Y, Si H, Niranjan T, Gruenwald A, Ahn S, Kato H, Pullman J, Gessler M, Haase VH. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120:4040-4054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 302] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 8. | Zhu F, Li T, Qiu F, Fan J, Zhou Q, Ding X, Nie J, Yu X. Preventive effect of Notch signaling inhibition by a gamma-secretase inhibitor on peritoneal dialysis fluid-induced peritoneal fibrosis in rats. Am J Pathol. 2010;176:650-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Kavian N, Servettaz A, Mongaret C, Wang A, Nicco C, Chéreau C, Grange P, Vuiblet V, Birembaut P, Diebold MD. Targeting ADAM-17/notch signaling abrogates the development of systemic sclerosis in a murine model. Arthritis Rheum. 2010;62:3477-3487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Nyhan KC, Faherty N, Murray G, Cooey LB, Godson C, Crean JK, Brazil DP. Jagged/Notch signalling is required for a subset of TGFβ1 responses in human kidney epithelial cells. Biochim Biophys Acta. 2010;1803:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Dees C, Zerr P, Tomcik M, Beyer C, Horn A, Akhmetshina A, Palumbo K, Reich N, Zwerina J, Sticherling M. Inhibition of Notch signaling prevents experimental fibrosis and induces regression of established fibrosis. Arthritis Rheum. 2011;63:1396-1404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 12. | Sawitza I, Kordes C, Reister S, Häussinger D. The niche of stellate cells within rat liver. Hepatology. 2009;50:1617-1624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Ono Y, Sensui H, Okutsu S, Nagatomi R. Notch2 negatively regulates myofibroblastic differentiation of myoblasts. J Cell Physiol. 2007;210:358-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 14. | Bolós V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 398] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 15. | Hansson EM, Lendahl U, Chapman G. Notch signaling in development and disease. Semin Cancer Biol. 2004;14:320-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1115] [Cited by in RCA: 1156] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 17. | Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609-2622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 481] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 18. | Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080-6089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 173] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 19. | Kokubo H, Lun Y, Johnson RL. Identification and expression of a novel family of bHLH cDNAs related to Drosophila hairy and enhancer of split. Biochem Biophys Res Commun. 1999;260:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 116] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Leimeister C, Schumacher N, Steidl C, Gessler M. Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. Mech Dev. 2000;98:175-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Purow B. Notch inhibitors as a new tool in the war on cancer: a pathway to watch. Curr Pharm Biotechnol. 2009;10:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124-5131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 307] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 23. | Fleisher AS, Raman R, Siemers ER, Becerra L, Clark CM, Dean RA, Farlow MR, Galvin JE, Peskind ER, Quinn JF. Phase 2 safety trial targeting amyloid beta production with a gamma-secretase inhibitor in Alzheimer disease. Arch Neurol. 2008;65:1031-1038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 250] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 24. | Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, Tsaparikos K, Jani JP, Hosea N, Sands M. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618-1628. [PubMed] |