Published online Jul 28, 2011. doi: 10.3748/wjg.v17.i28.3353
Revised: September 9, 2010
Accepted: September 16, 2010
Published online: July 28, 2011
AIM: To study Hepatitis B virus (HBV) infection and its association with hepatocellular carcinoma (HCC) at the miRNA level.
METHODS: Three cellular models were used to investigate miRNA expression changes during HBV infection: human HepG2 hepatoblastoma cell line as a model without HBV infection; HepG2 cell line transfected with a 1.3-fold full-length HBV genome as an acute infection model; and HepG2.2.15 cell line, which is derived from HepG2 and stably transfected with a complete HBV genome, as a chronic infection model. The miRNA levels were examined using microarray technology. To explore the relationship between HBV infection and HCC genesis at the miRNA level, we downloaded from national center for biotechnology information Gene Expression Omnibus an miRNA expression dataset derived from HCC patients, most of whom are HBV carriers. We compared the miRNA expression alterations during HBV infection with those in HCC patients, by analyzing miRNA expression change profiles statistically.
RESULTS: Seventy-seven and 48 miRNAs were differentially expressed during acute and chronic HBV infection, respectively. Among these miRNAs, 25 were in common, the intersection of which was significant under the hypergeometric test (P = 1.3 × 10-11). Fourteen miRNAs were observed to change coherently in the acute and chronic infections, with one upregulated and 13 downregulated. Eleven showed inverse changes during the two phases of infection; downregulated in the acute infection and upregulated in the chronic infection. The results imply that common and specific mechanisms exist at the miRNA level during acute and chronic HBV infection. Besides, comparative analysis of the miRNA expression changes during HBV infection with those in HCC indicates that, although miRNA expression changes during HBV infection are distinct from those in HCC patients (P < 2.2 × 10-16), they exhibited significant correlations (P = 0.0229 for acute infection; P = 0.0084 for chronic infection). Perturbation of miRNA expression during chronic HBV infection was closer to that in HCC patients than that during acute HBV infection. This observation implies the contribution of miRNAs to HCC genesis from HBV infection. According to their patterns of differential expression in acute and chronic HBV infection, as well as in HCC, miRNAs of potential research interest could be identified, such as miR-18a/miR-18b, miR-106a, miR-221 and miR-101. For instance, the gradient expression alteration of miR-221 in the above three phases, which is downregulated in acute HBV infection, normally expressed in chronic HBV infection, and upregulated in HCC, indicates that it may be a key effector for progression of the disease.
CONCLUSION: Our analysis provides insights into HBV infection and related HCC in relation to miRNAs, and reveals some candidate miRNAs for future studies.
- Citation: Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW, Huang AL, Liang Z. Hepatitis B virus and hepatocellular carcinoma at the miRNA level. World J Gastroenterol 2011; 17(28): 3353-3358
- URL: https://www.wjgnet.com/1007-9327/full/v17/i28/3353.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i28.3353
Hepatitis B virus (HBV) is a hepatotropic non-cytopathic DNA virus that belongs to the Hepadnaviridae family. It is a major cause of acute and chronic infections of the liver and can lead to hepatitis, cirrhosis and hepatocellular carcinoma (HCC)[1]. Many efforts have been made to investigate the liver diseases caused by HBV. However, attempts at treatment of chronic infections have had only limited success[2,3]. Due to the exclusive dependence of HBV on the host cellular machinery for its propagation and survival, investigation of the interactions between HBV and the host cell is crucial for the understanding of viral pathogenesis and development of novel antiviral therapies. Although the gene regulatory mechanisms involving host and viral proteins have been extensively explored, studies on miRNA-mediated regulation in viral infections are just emerging
As a class of small RNA molecules ~22 nucleotides in length, miRNAs have recently gained widespread attention as crucial regulators in complex gene regulatory networks[4-6]. miRNAs regulate gene expression by base-paring with the 3’ untranslated region (3’ UTR) of target mRNAs, which leads to mRNA degradation or translational silencing. Recent reports on the interactions between the host and the pathogen in viral infection through miRNAs shed light on the role of miRNAs as crucial effectors in the intricate host-pathogen interaction networks. Lecellier et al[7]. demonstrated for the first time that a mammalian miRNA, mir-32, restricts accumulation of the retrovirus primate foamy virus type 1 (PFV-1) in human cells Hariharan et al[8] have predicted that five human-encoded miRNAs potentially target the entire repertoire of accessory genes in HIV. Jopling et al[9] have reported an interesting case in which a liver-specific miRNA, mir-122, causes viral RNA accumulation of hepatitis C virus (HCV), by binding to the 5’ non-coding region of the viral genome. Pedersen and colleagues have shown that the antiviral effects of interferon (IFN)-β against HCV can be at least partially explained by miRNA-mediated modulation, and IFN-β treatment leads to a significant reduction in miR-122 expression, and induces miRNAs with sequence-predicted targets within the HCV genomic RNA[10]. To counteract the small-RNA-mediated interference, viruses have evolved to express suppressors that interfere with miRNA and siRNA pathways[7,11]. Several studies have shown that viruses also encode miRNAs, which can modulate cellular processes as well as regulate themselves[12-14]. Computational analysis has indicated the miRNA-encoding potential of HBV[15].
Previous works have pointed to the possibility of crosstalk between HBV and its host at the miRNA level. In the present study, we investigated human miRNAs that may be involved in acute and chronic HBV infection, via microarray profiling. We found that a significant number of differentially expressed miRNAs during acute and chronic infection overlapped, which were either coherently or inversely changed in the two phases of infection. This indicates that the two processes are both associated and different at the miRNA level. In addition, we explored the relationship between HBV infection and HCC genesis, by integrating our HBV infection dataset with a public miRNA expression dataset derived from a group of HCC patients, most of whom were HBV positive[16]. Our analysis demonstrated that perturbations of miRNA expression during HBV infection were significantly correlated with those in HCC, although they seemed distinct. Compared with acute HBV infection, chronic infection showed more consistent miRNA expression alterations with respect to HCC. In spite of this, there is a long way to go from the miRNA expression states during HBV infection to those that occur in HCC. The results implie that interference therapy at the miRNA level may provide a strategy to control the progression of serious liver diseases in HBV carriers.
Three cell models were used in this study: human HepG2 hepatoblastoma cell line as a model without virus infection; HepG2 cell line transfected with a 1.3-fold full-length HBV genome as an acute infection model; and HepG2.2.15 cell line, which is derived from HepG2 and stably transfected with a complete HBV genome, as a chronic infection model.
MiRNA expression profiles were determined at CapitalBio Corp (Beijing, China; http://www.capitalbio.com) by using mammalian miRNA arrays (version 3.0) which were designed based on the miRBase release 10.0 and contained 924 probes from humans, mice and rats. The arrays were scanned using a LuxScanTM laser confocal scanner and the images obtained were analyzed using LuxScan 3.0TM image analysis package. The raw miRNA expression data were quantile-normalized to correct for between-sample variations. A miRNA was determined as differentially expressed if its expression change was more than 1.5-fold, and it was identified as significantly changed using the Significance Analysis of Microarrays method with FDR < 0.05.
We downloaded the miRNA expression profiles of 78 HCC patients from Gene Expression Omnibus (GEO, GSE10694)[16]. For each patient, the expression data for the liver cancer tissue and the corresponding non-cancerous tissue were available. The microarray analysis was performed using a customized miRNA array produced by CapitalBio Corp, which was designed based on miRBase release 7.0 and contained 435 probes for human mature miRNAs, including 122 predicted ones. The data were normalized, and differentially expressed miRNAs identified using the same criteria as described above.
We generated miRNA expression change profiles (miECPs) for acute and chronic HBV infection by comparing the mean miRNA expression levels in the corresponding conditions to those in the uninfected model. Similarly, for the HCC patients, miECPs were obtained by dividing the miRNA expression levels in the patients’ non-cancerous tissues into those in the matched liver cancer tissues. Hierarchical clustering of the miECPs was performed to give a qualitative evaluation of the relationship between HBV infection and HCC. The significance of the observed deviations of HBV infection from HCC was determined by comparing the distributions of Pearson correlation coefficients (PCCs) between the miECPs of HBV infection and those of HCC, with the distribution of PCCs in HCC using the Kolmogorov-Smirnov test.
To investigate the association of miRNA expression alterations between HBV infection and HCC, we tested the null hypothesis that the correlation between the HBV infection miECPs and those for HCC was as weak as that between the random miECPs and HCC, which meant that the changes in miRNA expression during HBV infection were irrelevant to those observed in HCC. We first generated 10 000 random miECPs by sampling from the elements of the real miECPs. We then computed, for each random miECP, the median of its PCCs with the HCC miECPs. By pooling together the PCC medians of all the random miECPs, we constructed the null distribution for statistical analysis. The correlations between HBV infection miECPs and those for HCC, which were represented by their PCC medians, were evaluated by computing a P value, determined as the number of times random PCC medians were larger (one-tailed test for a positive PCC median) or smaller (one-tailed test for a negative PCC median) than the observed one.
We determined the global miRNA expression profiles elicited in the uninfected, acute infection and chronic infection models using CapitalBio mammalian miRNA arrays. Among the 570 human mature miRNAs investigated by the arrays, 77 and 48 were differentially expressed during acute and chronic infection, respectively, with respect to the uninfected model (Table 1). The former consisted of 21 upregulated and 56 downregulated miRNAs, and the latter contained 27 upregulated and 21 downregulated miRNAs. Among these differentially expressed miRNAs, 25 were in common between acute and chronic infection (Table 2); the intersection of which was significant by the hypergeometric test (P = 1.3 × 10-11). Fourteen of these miRNAs were observed to change coherently in acute and chronic infection, with one upregulated and 13 downregulated. Eleven were downregulated in acute infection, but inversely changed in chronic infection.
| Acute infection | Chronic infection | |
| Up | MiR-129-3p, miR-133a, miR-196b, miR-223, miR-296-5p, miR-302c, miR-361-3p, miR-365, miR-372, miR-409-3p, miR-518b, miR-562, miR-564, miR-574-3p, miR-612, miR-638, miR-634, miR-659, miR-663, miR-665, miR-940 | Let-7a, let-7b, let-7d, let-7g, let-7i, miR-23a, miR-25, miR-27a, miR-27b, miR-29a, miR-29b, miR-103, miR-146a, miR-146b-5p, miR-181a, miR-181b, miR-181c, miR-181d, miR-182, miR-196b, miR-222, miR-331-3p, miR-499-3p, miR-499-5p, miR-501-3p, miR-660, miR-888 |
| Down | Let-7a, let-7g, miR-15a, miR-15b, miR-16, miR-17, miR-18a, miR-19a, miR-19b, miR-20a, miR-20b, miR-23a, miR-25, miR-26a, miR-26b, miR-27a, miR-27b, miR-29b, miR-29c, miR-30a, miR-30b, miR-30c, miR-30e, miR-32, miR-34a, miR-92b, miR-96, miR-101, miR-103, miR-106a, miR-107, miR-122, miR-128a, miR-129-5p, miR-130a, miR-140-5p, miR-141, miR-146a, miR-146b-5p, miR-148a, miR-181a, miR-186, miR-192, miR-199a-5p, miR-200a, miR-200b, miR-215, miR-217, miR-221, miR-224, miR-301a, miR-338-3p, miR-374b, miR-454, miR-611, miR-923 | MiR-17, miR-18a, miR-18b, miR-19a, miR-19b, miR-20a, miR-20b, miR-26b, miR-92a, miR-92b, miR-101, miR-106a, miR-130a, miR-143, miR-148a, miR-193b, miR-199b-5p, miR-325, miR-338-3p, miR-378, miR-483-5p |
| Acute | ||
| Up | Down | |
| Chronic | ||
| Up | MiR-196b | Let-7a, let-7g, miR-23a, miR-25, miR-27a, miR-27b, miR-29b, miR-103, miR-146a, miR-146b-5p, miR-181a |
| Down | - | MiR-17, miR-18a, miR-19a, miR-19b, miR-20a, miR-20b, miR-26b, miR-92b, miR-101, miR-106a, miR-130a, miR-148a, miR-338-3p |
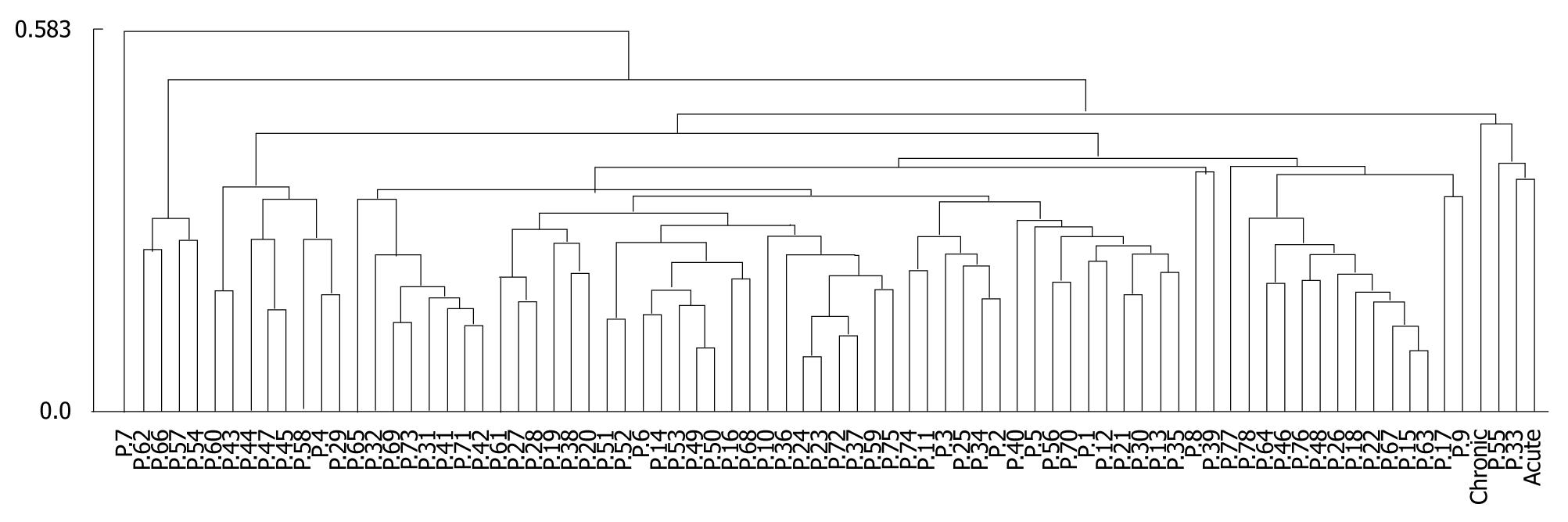
The close pathological associations between HBV infection and HCC genesis have been demonstrated by previous studies. It is of great interest to investigate the relationship between the two at the miRNA level. Recently, Li et al[16] performed an miRNA expression profiling analysis on a group of 78 HCC patients, 62 of whom were HBV carriers. This dataset provided us an opportunity to explore the problem of whether HBV infection exhibits similar miRNA expression changes as those observed in HCC. We identified 217 common miRNAs between our HBV infection dataset and that of Li et al. To summarize the expression changes of these miRNAs in HBV infection, we generated miECPs by taking ratios of the mean miRNA expression levels of acute and chronic infections, with respect to those of the uninfected model. Similarly, miECPs for HCC were generated by comparing the miRNA expression levels in liver cancer tissues against those in matched non-cancerous tissues. Hierarchical clustering of the miECPs indicated that alterations of miRNA expression were different for HBV infection and HCC, and more consistent alterations were observed for HCC (Figures 1). Although the visualization showed the separation of HBV infection from most HCC, however, several patients showed a more extreme distribution. Therefore, we asked whether the observed differences between HBV infection and HCC were significant.
Statistically, it was equivalent to the question of whether the miECPs of HBV infection were from the population represented by the HCC miECPs. To investigate this problem, we used a PCC to quantify the similarity between a pair of miECPs, and tested whether the distribution of PCCs between the miECPs of acute/chronic HBV infection and those from HCC were different from that between the HCC miECPs themselves. It was found that these two kinds of PCC distributions had a significant difference (Kolmogorov-Smirnov test, P < 2.2 × 10-16), and the latter had a significantly high median PCC of 0.26 compared with -0.07 (acute infection) and 0.09 (chronic infection) of the former. This demonstrates that the miRNA expression changes during HBV infection are distinct from those in HCC.
A PCC of 0 indicates linear independence, therefore, we were curious as to whether the observed small PCCs suggested that the miRNA expression alterations between HBV infection and HCC were unrelated. We reasoned that if the extent of correlation between the HBV infection miECPs and those of HCC were indistinguishable from that between the HCC miECPs and random ones, which are obviously irrelevant, they should be deemed as unrelated. The analysis demonstrated that the miECPs of acute HBV infection exhibited a significant negative correlation with those of HCC, with respect to the irrelevant random background (P = 0.0229); whereas the miECPs of chronic HBV infection showed a significant positive correlation (P = 0.0084). These results indicate that, although subtle, the miRNA expression changes during HBV infection were correlated with those observed in HCC. Compared to acute HBV infection, chronic infection was closer to HCC at the miRNA level
Using the same criteria as for the analysis of the HBV infection dataset, we identified from the HCC dataset 46 differentially expressed miRNAs in liver cancer tissues, with respect to the corresponding non-cancerous tissues. The intersection of differentially expressed miRNAs between HBV infection and HCC is summarized in Table 3. Eight of the 10 differentially expressed miRNAs common to the acute HBV infection and HCC datasets were inversely changed, whereas only three of the eight differentially expressed miRNAs common to the chronic HBV infection and HCC datasets exhibited opposite alterations. The observation also indicates that chronic HBV infection has closer relationship with HCC than acute infection does. According to their patterns of differential expression in acute and chronic HBV infection, as well as in HCC, miRNAs with potential research interest could be identified.
| Acute | Chronic | |||
| Up | Down | Up | Down | |
| HCC | ||||
| Up | - | MiR-15b | MiR-25 | MiR-18a MiR-18b MiR-106a |
| MiR-18a | MiR-103 | |||
| MiR-25 | MiR-182 | |||
| MiR-103 | MiR-222 | |||
| MiR-106a | ||||
| MiR-107 MiR-221 | ||||
| MiR-224 | ||||
| Down | - | MiR-101 MiR-29c | - | MiR-101 |
Our microarray profiling analysis of miRNAs during HBV infection showed that a significant number of miRNAs differentially expressed during acute and chronic infection were overlapped, which were either coherently or inversely changed in the two phases of infection. These observations indicate that common and phase-specific mechanisms for acute and chronic infection may exist at the miRNA level. To examine the reliability of our expression dataset, we compared our results with a recent study performed by Liu et al[17], who investigated the miRNA expression alterations in the HepG2.2.15 cell line with respect to HepG2. They used an early version of CapitalBio mammalian miRNA arrays, which contains probes for 435 human mature miRNAs, a subset of the miRNAs present on our arrays. Similar to our analysis, Liu et al used SAM and fold changes to determine differentially expressed miRNAs. However, they required a larger extent of expression alteration of at least three fold, compared with our 1.5. Under their criteria, eleven and seven miRNAs present on the arrays were identified as upregulated and downregulated, respectively. Between our chronic infection data and theirs, seven upregulated (miR-23a, miR-146a, miR-181a, miR-181b, miR-181c, miR-181d and miR-196b) and three downregulated (miR-17, miR-338-3p and miR-378) miRNAs were in common. The intersection was significant by the hypergeometric test (P = 4.45 × 10-7 for the upregulated case; P = 3 × 10-3 for the downregulated case), which demonstrated the reproducibility of our miRNA profiling experiments.
By integrating a miRNA expression dataset of HCC patients[16], most of whom were HBV positive, we were able to investigate the relationship between HBV infection and HCC at the miRNA level, and also had an opportunity to explore miRNA expression changes with the progression of disease, namely from acute HBV infection to chronic infection and at last to HCC status. The statistical analysis of miRNA expression change profiles suggested that the perturbation of miRNA expression during HBV infection is different from that in HCC, however, they are correlated. The pattern of miRNA expression during different phases of disease offers a means to discover miRNAs that might be of great importance for further research. It could be conjectured that miRNAs that exhibit opposite alterations between HBV infection and HCC may be of importance for the entry into a serious disease state, for instance, miR-18a/miR-18b and miR-106a[18], which have already been reported to be involved in HCC. Those showing a gradient alteration may be key effectors for the progression of disease. For example, miR-221, which is downregulated in acute HBV infection, and normally expressed in chronic HBV infection and upregulated in HCC, has been shown recently to contribute to liver tumorigenesis[19]. Also, miRNAs showing coherent alteration patterns among the disease states may be related to the establishment and maintenance of these disease states. For instance, aberrant expression of miR-101, which remains downregulated in acute and chronic HBV infection, as well as in HCC, is closely associated with HCC development[20,21].
In summary, the correlation between HBV infection and HCC can be identified at the miRNA level. Compared with acute HBV infection, miRNA expression changes during chronic HBV infection are closer to those in HCC. However, there is a long way to go from miRNA expression states of HBV infection to those of HCC, which may provide us with opportunities to control the progression of serious liver diseases via interference therapy at the miRNA level.
Hepatitis B virus (HBV) is a major cause of liver infection and severe liver diseases. The survival and propagation of HBV exclusively depends on the host cellular machinery, therefore, investigation of HBV-host interactions is crucial for understanding viral pathogenesis and development of antiviral therapies.
Although the gene regulatory mechanisms involving host and viral proteins have been extensively explored, studies on miRNA-mediated regulation in viral infections are just emerging. Recent works on the pathogen-host interactions through miRNAs point to the possibility of crosstalk between HBV and the host at the miRNA level.
The present study indicates that acute and chronic HBV infections are both associated and different at the miRNA level. In addition, the perturbations of miRNA expression during HBV infection are significantly correlated with those in hepatocellular carcinoma (HCC), although they seem distinct. In spite of this, there is a long way to go from miRNA expression states during HBV infection to those that occur in HCC.
The study implies that interference therapy at the miRNA level may provide a strategy to control the progression of serious liver diseases in HBV carriers and reveals some candidate miRNAs for future studies.
miRNAs are a class of small RNA molecules, ~22 nucleotides in length, which regulate gene expression by base-paring with the 3’ untranslated region of target mRNAs, leading to mRNA degradation or translational silencing. miRNAs have recently gained widespread attention as crucial regulators in complex gene regulatory networks.
This study illustrated that the miRNA level is correlated in HBV infection and HCC. miRNA expression levels during HBV acute and chronic infections were investigated in cell lines, and the clustering between their profiles and those in HCC showed some insights into associations between HBV infection and HCC.
Peer reviewer: Dae-Yeul Yu, PhD, Professor, AgingResearch Center, Korea Research Institute of Bioscience and Biotechnology, 111 Gwahangno, Yuseong-gu, Daejeon 305-806, Korea
S- Editor Sun H L- Editor Kerr C E- Editor Ma WH
| 1. | Seeger C, Mason WS. Hepatitis B virus biology. Micro biol Mol Biol Rev. 2000;64:51-68. |
| 2. | Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662. |
| 3. | Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347-356. |
| 5. | Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641-26645. |
| 7. | Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557-560. |
| 8. | Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214-1218. |
| 9. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. |
| 10. | Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922. |
| 11. | Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768-2780. |
| 12. | Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682-686. |
| 13. | Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269-276. |
| 14. | Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499-5508. |
| 15. | Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124-126. |
| 16. | Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616-1622. |
| 17. | Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl). 2009;122:10-14. |
| 18. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and no n-tumorous tissues. Oncogene. 2006;25:2537-2545. |
| 19. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. |
| 20. | Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J, Sun Z, Wei L, Zheng X. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194-1220. |
| 21. | Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142. |