Published online Sep 21, 2010. doi: 10.3748/wjg.v16.i35.4460
Revised: May 18, 2010
Accepted: May 25, 2010
Published online: September 21, 2010
AIM: To evaluate the presence and cross-reactive antibodies against hypervariable region 1 (HVR1) in hepatitis C virus (HCV) infected patients and its relationship with the progression of the disease.
METHODS: Sixteen representative HVR1 proteins selected from a unique set of 1600 natural sequences were used to semiquantitate the cross-reactivity of HVR1 antibodies in the sera of HCV patients. Fifty-five chronic HCV patients including 23 with asymptomatic mild hepatitis, 18 with chronic hepatitis and 16 with liver cirrhosis patients were studied.
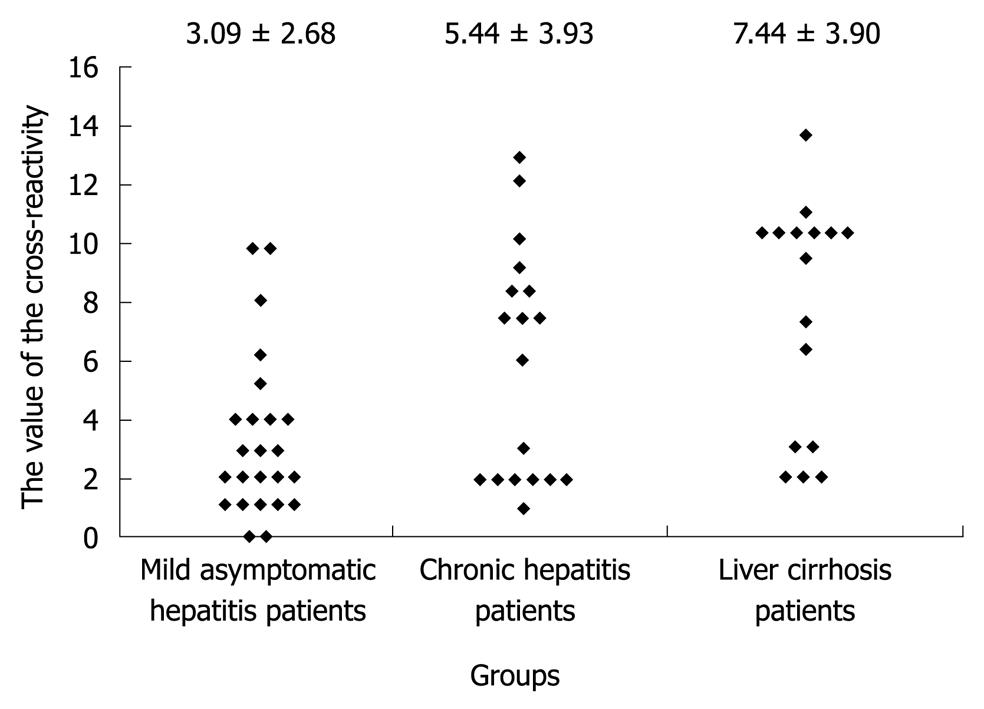
RESULTS: The degree of the cross-reactivity of anti-HVR1 antibodies in 23 patients with mild asymptomatic hepatitis was 3.09 ± 2.68, which was significantly lower than in those with chronic hepatitis (5.44 ± 3.93, P < 0.05) and liver cirrhosis (7.44 ± 3.90, P < 0.01). No correlation was observed between the broadness of the cross-reactivity anti-HVR1 antibodies and patient’s age, infection time, serum alanine aminotransferase activity, or serum HCV-RNA concentration. It was the breath of cross-reactivity rather than the presence of anti-HVR1 antibody in HCV sera that was associated with the progression of liver disease.
CONCLUSION: The broadly cross-reactive HVR1 antibodies generated in natural HCV patients can not neutralize the virus, which results in persistent infection in patients with chronic hepatitis.
- Citation: Xiu BS, Feng XY, He J, Wang GH, Zhang XY, Zhang HQ, Song XG, Chen K, Ling SG, Zhu CX, Wei L, Rao HY. Evaluation of cross-reactive antibody response to HVR1 in chronic hepatitis C. World J Gastroenterol 2010; 16(35): 4460-4466
- URL: https://www.wjgnet.com/1007-9327/full/v16/i35/4460.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i35.4460
Hepatitis C virus (HCV) is the major causative agent of post-transfusional and sporadic non-A, non-B hepatitis. HCV infection is persistent in over 70% of cases and may result in various forms of chronic hepatitis and other liver diseases, ranging from asymptomatic mild chronic hepatitis to cirrhosis and hepatocellular carcinoma[1].
HCV is a RNA virus that replicates with a high rate of mutation. The maximal variation is confined to a short sequence of the second envelope glycoprotein (E2), and has been designated the hypervariable region 1 (HVR1)[2]. It is accepted that the immune escape is the main mechanisms responsible for HCV persistence. It indicated that the viral and humoral responses may be two important factors in pathogenesis of hepatitis C[3,4]. The virological studies reveal that HCV circulates within an infected host as a heterogeneous viral population containing genetically distinct, but closely related variants, known as quasispecies[5,6]. The diversity of quasispecies and its relationship with the progression of liver disease and interferon treatment were largely studied by single-strand conformation polymorphism or sequencing of HVR1[7-9]. However, for the high antigenic variability of HVR1, the humoral response against the HVR1 could not be easily characterized in chronic HCV infection.
Early observations have suggested that the HVR1 is a critical neutralization domain. Antibodies to HVR1 in human serum have been shown to block viral attachment and protect chimpanzees from HCV infection[10,11]. The recent development of model systems, including retroviral HCV pseudotypes (HCVpp) and recombinant cell-culture derived infectious virions (HCVcc), has established the neutralization of HVR1 antibodies[12]. In viral infections, the appearance of neutralizing antibody is usually a prognostic marker which coincides with the onset of recovery from the disease and viral elimination from the circulation. Although an early anti-HVR1 response is associated with self-limiting acute infection[13], anti-HVR1 antibodies are frequently produced in the majority of chronically infected individuals and appear to coexist with the HVR1 variants[14]. In most instances, the sera of HCV infected patients are frequently cross-reactive with unrelated HVR1 sequences[15,16]. However, the cross-reactive nature of anti-HVR1 responses and the relationship between the cross-reactivity and the progression of hepatitis are largely unknown.
The HCVpp and HCVcc could characterize the neutralizing antibodies in HCV infection, but could not be used to detect antibodies to the hypervariable region conveniently. In this report, we used a protein microarray immobilized with a set of HVR1 proteins selected by alignments to study the humoral anti-HVR1 response in 57 cases of chronic HCV infections with different clinical course. Our findings suggest that the broadly cross-reactive antibody responses against HVR1 are associated with the progression of liver disease with chronic HCV infection, and this may contribute to the better understanding of HCV natural infection and its prognosis.
All of the patients from the rural area of Zhao County in Hebei Province with a history of plasmapheresis before 1990 were first diagnosed as having HCV infection in 1993. Fifty-seven of them were followed up till 2007 and were seropositive for anti-HCV antibodies by a third-generation enzyme-linked immunosorbent assay (ELISA) technique (HCV ELISA 3.0, Ortho Diagnostic Systems, Raritan, NJ). Serum HCV-RNA was measured by quantitative completive reverse transcription polymerase chain reaction analysis (Amplicor, Roche Diagnostic Systems, Inc., Branchburg, NJ) according to the manufacturer’s protocol. According to the results of ultrasound (US) evaluation and clinical symptoms, 23 of them had asymptomatic hepatitis, 18 chronic hepatitis and 16 liver cirrhosis. The US parameters, including the assessment of liver surface, liver parenchyma, hepatic vessel, spleen index, were used to define the severity of the liver disease[17,18]. The clinical symptoms, including fatigue, nausea, abdominal, pain, anorexia, itching, dark urine and extra hepatic manifestations, were used to define the asymptomatic and chronic hepatitis. The duration of the infection was calculated from the time of plasmapheresis. No antiviral treatment was given until blood sampling, and none of them was infected with hepatitis A or B viruses, and human immunodeficiency virus. The study protocol was approved by the committee of ethics of the authors’ institution.
The 1600 HVR1 sequences from Genbank were ranked according to the results of multiple sequence alignments using Biosun molecular biology software[19,20]. The representative HVR1 sequence with properties of highest similarity in each group was selected according to their pairs (SP) scores.
The representative HVR1 sequences were modified considering the Escherichia coli (E. coli) favorable codon usage. The full-length genes were synthesized chemically and cloned into the BamHI-EcoRI sites of the expression vector pGEX4T-2. All the genes were expressed as fusion protein with glutathione transferases (GST) in E. coli. Then the recombinant HVR1 antigens were purified by GST resin.
Microplates were coated with 0.3 μg recombinant HVR1 antigen in 100 mmol/L phosphate buffer (pH 7.4) by incubation overnight at 4°C. The plates were blocked with the phosphate buffer containing 0.2% BSA at 4°C for 3 h, and incubated with 100 μL of the serum sample 1:10 diluted with a sample buffer (100 mmol/L sodium phosphate buffer pH 7.5 containing 10% goat serum and 0.05% Tween) at 37°C for 1 h. After being washed for five times with 100 mmol/L sodium phosphate buffer (pH 7.5) containing 0.05% Tween, the plate was incubated for 30 min at 37°C with 1:25 000 diluted horseradish peroxidase-conjugated monoclonal antibody against human IgG. After washing, the reaction was visualized in the substrate buffer (50 mmol/L sodium phosphate-citric acid buffer, pH 5.0, containing 0.4 mg/mL tetramethylbenzidine and 0.4 μL/mL of 30% hydrogen peroxide). The reaction was stopped by adding 50 μL of 2 mol/L sulfuric acid, and the absorbance was measured in a microplate ELISA reader at 450 nm.
All of the recombinant HVR1 antigens were diluted in spotting buffer (CapitalBio Corporation), and were printed onto the silylanized slides (CapitalBio Corporation) by a SmartArrayer™ 48 microarray printer. The microarray included all of selected HVR1 antigens printed in three replicates with IL-1 and human IgG as negative and positive control, respectively. The arrays were blocked with blocking buffer [phosphate buffered saline (PBS) containing 2% BSA] at room temperature for 3 h, and were air dried.
The arrays were covered with the sera of HCV infected patients at 1/10 in the blocking buffer for 40 min at 37°C and washed 3 times with PBS Tween-20 (PBST) solution dissolved in PBS to a final concentration of 0.05%. After rinsed twice with PBS, the slides were incubated in Cy3 conjugated protein A for 30 min at 37°C and washed 3 times with PBST solution. The slides were finally air dried under short centrifugation and examined in a LuxScan™ 10K microarray scanner (CapitalBio Corporation). The fluorescence intensities were determined by the software taken by the scanner.
The number of HVR1 antigen reacted with the HCV sera on the chip was used to define the range of cross-reactive responses for each sample. Qualitative data were analyzed using the χ2 test or the Fisher’s exact test when necessary. Quantitative values were compared using the Student’s t test, and the Kruska Wallis test when necessary. P values lower than 0.05 were considered significant. All statistical calculations were performed using the SPSS for Windows, version 6.0 software package.
A total of 1600 HVR1 sequences were collected from Genebank to construct the database by Biosun software. The duplicated sequences were removed from the database to obtain a unique set of 843 natural HVR1 sequences. Thirty HVR1 sequences were selected from the database according to the results of multiple sequence alignment using Biosun software. All were cloned and expressed in E. coli, and the 30 HVR1 was successively selected according to their immunological reactivity tested by ELISA.
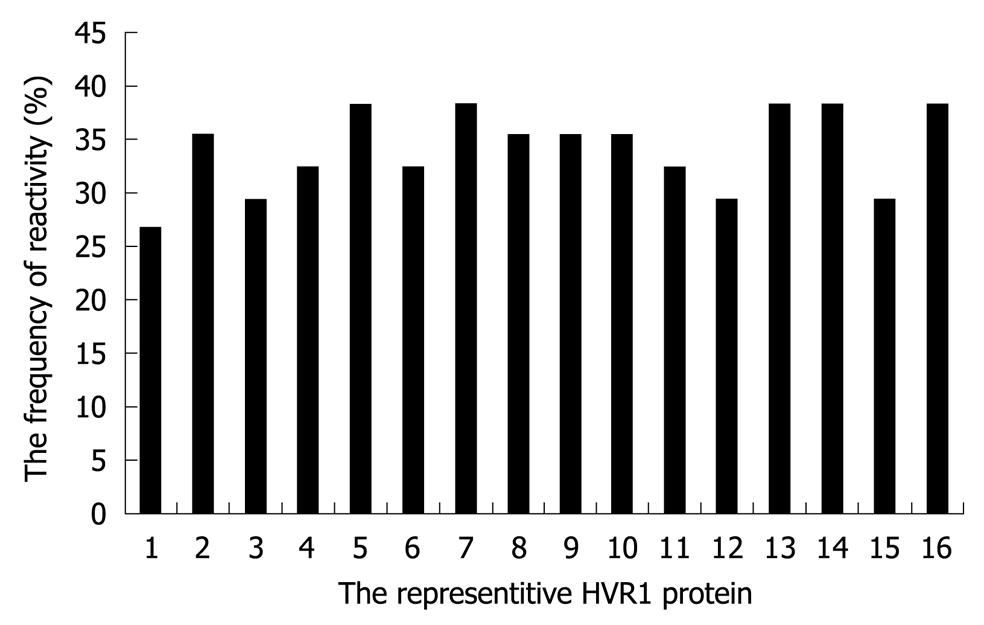
To evaluate the cross-reactivity of HCV sera, we need the representative HVR1 sequences which display not only the sequence diversity but also immunological variability. The non-overlapping reactivity of the representative HVR1 with the sera of the in-house panel was emphasized in this selection. As a result, 16 representative peptides (Figure 1) were selected from the 30 HVR1 with the cross-reactivity frequency ranging from 20% to 30% with the in-house panel (Figure 2). The 16 representative HVR1s could react with 32/34 sera in the panel and span the overall reactivity of the above 30 HVR1.
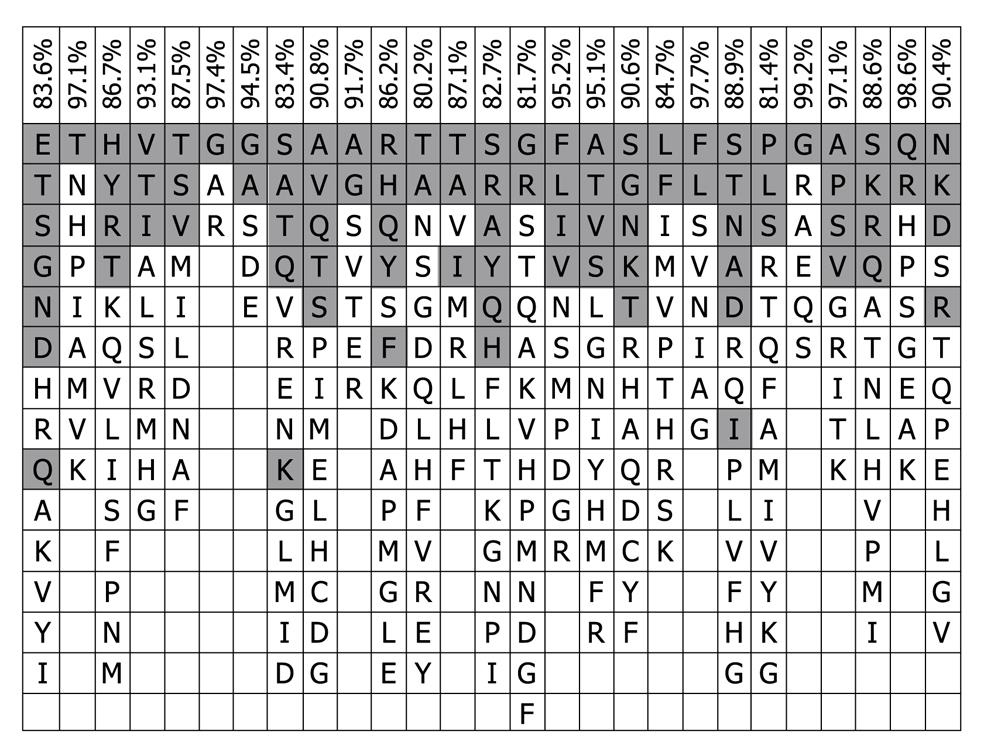
The 16 representative HVR1 sequences were aligned further as shown in Table 1 and compared with the consensus pattern of 843 HVR1 sequences. The residues of 16 representative HVR1 sequences accounted for 90.4% (median value) of the observed variability of the consensus pattern of 843 HVR1 sequences (Figure 3). This indicated that the 16 representative sequences we selected could cover most of the natural variability of HVR1 at least from the amino acid residue sequence.
| Percentages of amino acid sequence variation | |||||||||||||||
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
| 1 | 51.9 | 40.7 | 29.6 | 48.1 | 40.7 | 40.7 | 44.4 | 40.7 | 29.6 | 40.7 | 70.4 | 59.3 | 48.1 | 48.1 | 37.0 |
| 2 | - | 44.4 | 51.9 | 51.9 | 55.6 | 66.7 | 44.4 | 44.4 | 51.9 | 48.1 | 63.0 | 59.3 | 40.7 | 48.1 | 44.4 |
| 3 | - | 33.3 | 51.9 | 66.7 | 59.3 | 51.9 | 48.1 | 40.7 | 55.6 | 55.6 | 63.0 | 55.6 | 37.0 | 33.3 | |
| 4 | - | 44.4 | 48.1 | 37.0 | 51.9 | 37.0 | 44.4 | 48.1 | 66.7 | 59.3 | 44.4 | 51.9 | 44.4 | ||
| 5 | - | 44.4 | 70.4 | 37.0 | 33.3 | 40.7 | 51.9 | 59.3 | 59.3 | 33.3 | 55.6 | 44.4 | |||
| 6 | - | 59.3 | 55.6 | 40.7 | 51.9 | 51.9 | 74.0 | 59.3 | 40.7 | 66.7 | 51.9 | ||||
| 7 | - | 59.3 | 55.6 | 59.3 | 55.6 | 63.0 | 48.1 | 63.0 | 51.9 | 59.3 | |||||
| 8 | - | 48.1 | 40.7 | 59.3 | 63.0 | 55.6 | 44.4 | 40.7 | 48.1 | ||||||
| 9 | - | 51.9 | 66.7 | 44.4 | 48.1 | 29.6 | 51.9 | 51.9 | |||||||
| 10 | - | 48.1 | 51.9 | 66.7 | 48.1 | 48.1 | 51.9 | ||||||||
| 11 | - | 63.0 | 51.9 | 51.9 | 66.7 | 44.4 | |||||||||
| 12 | - | 66.7 | 66.7 | 66.7 | 55.6 | ||||||||||
| 13 | - | 55.6 | 63.0 | 66.7 | |||||||||||
| 14 | - | 44.4 | 51.9 | ||||||||||||
| 15 | - | 48.1 | |||||||||||||
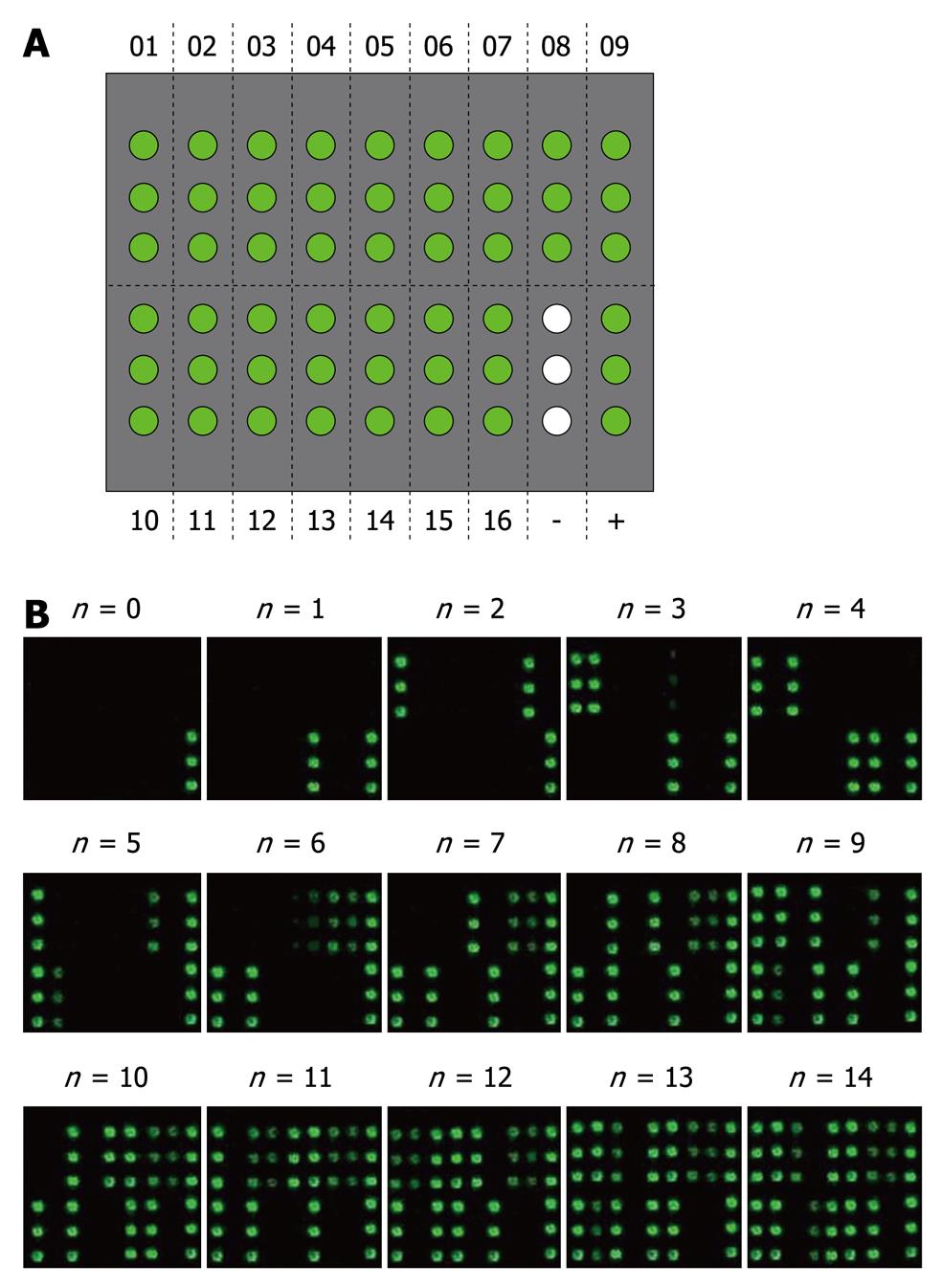
The microarray with 16 HVR1 immobilized as antigens was used to evaluate the cross-reactivity of anti-HVR1 antibodies, and GST and human IgG were used as negative and positive controls, respectively (Figure 4A). The value of the cross-reactive response for each sample was evaluated according to the number of HVR1 antigens reacted with the HCV sera (Figure 4B).
Considering the size of the sample, we devised a microarray to evaluate the broadness of serological reactivity to 16 HVR1 selected antigens. The basal features of patients and the results of the microarray are summarized in Table 2. The value of the cross-reactivity observed in individual cases did not correlate with patient’s age (r = 0.0063, P > 0.05), infection time (r = 0.14, P > 0.05), serum alanine aminotransferase activity (r = 0.181, P > 0.05), or serum HCV-RNA concentration (r = 0.125, P > 0.05). No differences related to sex were found (5.93 ± 4.18 in men and 4.7 ± 3.54 in women, P > 0.05).
| Mild (n = 23) | Moderate (n = 18) | Hepatic cirrhosis (n = 16) | |
| Age (yr) | 44.5 ± 7.3 | 47.2 ± 10.8 | 45.4 ± 8.0 |
| Time of infection | 16.2 ± 4.3 | 18.1 ± 4.3 | 17.8 ± 5.7 |
| Sex (M/F) | 9/14 | 7/11 | 11/5 |
| ALT | 42.3 ± 48.1 | 39.4 ± 43.2 | 94.1 ± 167.6 |
| HCV-RNA (copes × 103) | 2189 ± 4562 | 4153 ± 11564 | 1582 ± 1676 |
| Cross-reactivity | 3.09 ± 2.68ad | 5.44 ± 3.93 | 7.44 ± 3.90 |
| Positive for anti-HVR1 | 21 | 18 | 16 |
The degree of the cross-reactivity of anti-HVR1 antibodies (Figure 5) in 23 patients with mild chronic hepatitis was 3.09 ± 2.68, which was significantly different from that in those with severe hepatitis (5.44 ± 3.93, P < 0.05) and liver cirrhosis (7.44 ± 3.90, P < 0.01).
The appearance of HVR1 antibodies was defined positive when the serum could react with more than 1 HVR1 antigen (include 1). In the sera of 23 mild chronic patients, 21 were anti-HVR1 positive, and all were anti-HVR1 positive in moderate hepatitis and liver cirrhosis patients (Table 2). The appearance of HVR1 antibodies was found similar in the three groups of patients (P > 0.05).
It is well known that the HCV infection is persistent in up to 85% of cases and may result in mild chronic hepatitis, cirrhosis and hepatocellular carcinoma. There are no obvious serologic features that can work as a prognostic marker although Zibert et al[11] found that early appearance of anti-HVR1 antibodies within the first 6 mo is associated with self-limited HCV infections. In this study, all the patients were not at the early stage, and had the disease for at least 10 years. We found that anti-HVR1 antibodies were widely produced in chronic patients, and there was no significant difference in mild hepatitis, moderate hepatitis and liver cirrhosis. That means that the anti-HVR1 antibodies could not be used in prognostic and turnover studies of chronic HCV infection, which was coincided with other studies[14,21].
It has been reported that the anti-HVR1 antibodies in HCV infected individuals could react with more than one variant of HVR1[15,16,22]. In this study, 16 representative HVR1 antigens distributed homogeneously were used to evaluate the cross-reactivity of HVR1 antibodies. Our data suggest that the cross-reactivity of HVR1 antibodies in moderate hepatitis and liver cirrhosis is broader than that in mild chronic hepatitis. Mondelli et al[23] found that the heterogeneity of cross-reactive antibodies was significantly higher in patients with chronic hepatitis than in those with acute hepatitis. This suggested that it is the broadly cross-reactivity of HVR1 antibodies that were associated with the progression of liver disease and could be a new marker in prognostic study of chronic HCV infection, but not the appearance of anti-HVR1 antibodies.
In our studies, the patients selected owned similar background with plasmapheresis transmitted, genotype 1b,without any previous treatment and at the same geographical area. Considering the invasion and complications of the liver biopsy[24], US was used to assess the severity of hepatitis C. For the limitation of US determination, the patients were divided into asymptomatic hepatitis, chronic hepatitis and liver cirrhosis group[17,18]. To elevate the accuracy of the US, the patients with indeterminate US sign were excluded from our studies. In addition, the clinical symptoms were considered to assess the severity of hepatitis C. The patients with no more than one clinical symptom were defined as having the asymptomatic hepatitis. This shows that the chronic hepatitis group in our study consisted of moderate and severe liver diseases. Further studies will focus on the patients with liver biopsy to give detailed information about the relationship between the cross-reactivity antibodies of HVR1 with the severity of hepatitis C.
HCV exists in the bloodstream of infected individual as quasispecies, and quasispecies nature of HCV may confer important biological properties to the virus, including immunologic escape, viral persistence, and resistance to antiviral agents[25,26]. There was greater nucleotide sequence diversity between HCV variants in isolates from patients with more advanced liver diseases[27,28]. Our data indicate that the broadly cross-reactivity of anti-HVR1 antibodies was associated with the advanced liver disease, and was immune reaction responding to heterogeneity of the HCV quasispecies at certain time in chronic patients. Using the sequential serum samples and HCVpp model, HANA verified that the mutation of HVR1 resulted in loss of recognition of the cognate antibody response and escape from antibody-mediated neutralization in chronic patients[29]. It is the presence of the immune pressure and mutation of quasispecies which lead to the persistence of HCV.
It is mentioned that HVR1 domains of HCV appear to contain a neutralizing epitope[10-12]. It is important to acquire broadly cross-reactive HVR1 antibodies for the development of HCV vaccines[30]. Obviously, cross-reactive HVR1 antibodies in chronic patients failed to clear viral infection in our studies. Recently, interfering mechanism of neutralizing antibodies was established using two different antibodies against E1. That means that the presence of an abundance of neutralizing antibodies will interfere with the neutralizing activity to HCV[31]. The same mechanism may exist for the HVR1 antibodies.
With the 16 representative HVR1 antigens, we developed an assay to evaluate cross-reactivity antibodies against HVR1 and discussed the humoral anti-HVR1 response and its relationship with the severity of the liver disease with chronic HCV infection. Our study may contribute to the better understanding of the knowledge of immune response of natural HCV infection, which will benefit the study of HCV vaccines based on the HVR1 and HCV prognosis.
Although an early anti-hypervariable region 1 (HVR1) response is associated with self-limiting acute infection, anti-HVR1 antibodies are frequently produced in the majority of chronically infected individuals and appear to coexist with the HVR1 variants. In most instances, the sera of hepatitis C virus (HCV) infected patients are frequently cross-reactive with unrelated HVR1 sequences. However, the cross-reactive nature of anti-HVR1 responses and the relationship between the cross-reactivity and the progression of hepatitis are largely undefined.
HCV is a RNA virus that replicates with a high rate of mutation. The virological studies revealed that HCV circulates within an infected host as a heterogeneous viral population containing genetically distinct, but closely related variants, known as quasispecies. The diversity of quasispecies and its relationship with the progression of liver disease and interferon treatment were largely studied by single-strand conformation polymorphism or sequencing of HVR1. However, for the high antigenic variability of HVR1, the humoral response against the HVR1 could not be easily characterized in chronic HCV infection. HVR1 domains of HCV appear to contain a neutralizing epitope. To acquire broadly cross-reactive HVR1 antibodies is important for the development of HCV vaccine. Recently, interfering mechanism of neutralizing antibodies was established using two different antibodies against E1.
For the high antigenic variability of HVR1, the humoral response against the HVR1 could not be easily characterized in chronic HCV infection. In this report, the authors used a protein microarray immobilized with a set of HVR1 proteins selected by alignments to study the humoral anti-HVR1 response in chronic HCV infections with different clinical courses.
The findings suggest that the broadly cross-reactive antibodies response against HVR1 is associated with the progression of liver disease with chronic HCV infection, and this may contribute to the better understanding of HCV natural infection and its prognosis.
This is an interesting study designed to evaluate the presence of cross-reactive antibodies against the HVR1 in HCV infected patients and its relationship with disease progression.
Peer reviewers: Dr. BS Anand, Professor, Digestive Diseases Section (111D), VA Medical Center, 2002 Holcombe Blvd., Houston, TX 77030, United States; Seyed-Moayed Alavian, MD, Professor, Gastroenterology and Hepatology, Department of Internal Medicine, Baqiyatallah University of Medical Sciences and Tehran Hepatitis Center, PO Box 14155-3651-Tehran, Iran
S- Editor Wang YR L- Editor Ma JY E- Editor Zheng XM
| 2. | Weiner AJ, Geysen HM, Christopherson C, Hall JE, Mason TJ, Saracco G, Bonino F, Crawford K, Marion CD, Crawford KA. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468-3472. |
| 3. | Zhou D, Fan X, Tan D, Xu Y, Tavis JE, Di Bisceglie AM. Separation of near full-length hepatitis C virus quasispecies variants from a complex population. J Virol Methods. 2007;141:220-224. |
| 4. | Rothman AL, Morishima C, Bonkovsky HL, Polyak SJ, Ray R, Di Bisceglie AM, Lindsay KL, Malet PF, Chang M, Gretch DR. Associations among clinical, immunological, and viral quasispecies measurements in advanced chronic hepatitis C. Hepatology. 2005;41:617-625. |
| 5. | Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107-123. |
| 6. | Gao G, Stuver SO, Okayama A, Tsubouchi H, Mueller NE, Tabor E. The minimum number of clones necessary to sequence in order to obtain the maximum information about hepatitis C virus quasispecies: a comparison of subjects with and without liver cancer. J Viral Hepat. 2005;12:46-50. |
| 7. | Honda M, Kaneko S, Sakai A, Unoura M, Murakami S, Kobayashi K. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology. 1994;20:1144-1151. |
| 8. | Li H, McMahon BJ, McArdle S, Bruden D, Sullivan DG, Shelton D, Deubner H, Gretch DR. Hepatitis C virus envelope glycoprotein co-evolutionary dynamics during chronic hepatitis C. Virology. 2008;375:580-591. |
| 9. | Alexopoulou A, Dourakis SP. Genetic heterogeneity of hepatitis viruses and its clinical significance. Curr Drug Targets Inflamm. Allergy. 2005;4:47-55. |
| 10. | Rothman AL, Morishima C, Bonkovsky HL, Polyak SJ, Ray R, Di Bisceglie AM, Lindsay KL, Malet PF, Chang M, Gretch DR. Associations among clinical, immunological, and viral quasispecies measurements in advanced chronic hepatitis C. Hepatology. 2005;41:617-625. |
| 11. | Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology. 1997;25:1245-1249. |
| 12. | Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci USA. 2003;100:14199-14204. |
| 13. | Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653-661. |
| 14. | Mondelli MU, Cerino A, Segagni L, Meola A, Cividini A, Silini E, Nicosia A. Hypervariable region 1 of hepatitis C virus: immunological decoy or biologically relevant domain? Antiviral Res. 2001;52:153-159. |
| 15. | Scarselli E, Cerino A, Esposito G, Silini E, Mondelli MU, Traboni C. Occurrence of antibodies reactive with more than one variant of the putative envelope glycoprotein (gp70) hypervariable region 1 in viremic hepatitis C virus-infected patients. J Virol. 1995;69:4407-4412. |
| 16. | Hjalmarsson S, Blomberg J, Grillner L, Pipkorn R, Allander T. Sequence evolution and cross-reactive antibody responses to hypervariable region 1 in acute hepatitis C virus infection. J Med Virol. 2001;64:117-124. |
| 17. | Wang JH, Changchien CS, Hung CH, Eng HL, Tung WC, Kee KM, Chen CH, Hu TH, Lee CM, Lu SN. FibroScan and ultrasonography in the prediction of hepatic fibrosis in patients with chronic viral hepatitis. J Gastroenterol. 2009;44:439-446. |
| 18. | Colli A, Fraquelli M, Andreoletti M, Marino B, Zuccoli E, Conte D. Severe liver fibrosis or cirrhosis: accuracy of US for detection--analysis of 300 cases. Radiology. 2003;227:89-94. |
| 19. | Li W, Ying X. Mprobe 2.0: computer-aided probe design for oligonucleotide microarray. Appl Bioinformatics. 2006;5:181-186. |
| 20. | Xiu BS, Ling SG, Song XG, Zhang HQ, Chen K, Zhu CX. Cross-reactivity of hypervariable region 1 chimera of hepatitis C virus. World J Gastroenterol. 2003;9:1256-1260. |
| 21. | Isaguliants MG, Widell A, Zhang SM, Sidorchuk A, Levi M, Smirnov VD, Santantonio T, Diepolder HM, Pape GR, Nordenfelt E. Antibody responses against B-cell epitopes of the hypervariable region 1 of hepatitis C virus in self-limiting and chronic human hepatitis C followed-up using consensus peptides. J Med Virol. 2002;66:204-217. |
| 22. | Puntoriero G, Meola A, Lahm A, Zucchelli S, Ercole BB, Tafi R, Pezzanera M, Mondelli MU, Cortese R, Tramontano A. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 1998;17:3521-3533. |
| 23. | Mondelli MU, Cerino A, Lisa A, Brambilla S, Segagni L, Cividini A, Bissolati M, Missale G, Bellati G, Meola A. Antibody responses to hepatitis C virus hypervariable region 1: evidence for cross-reactivity and immune-mediated sequence variation. Hepatology. 1999;30:537-545. |
| 24. | Piccinino F, Sagnelli E, Pasquale G, Giusti G. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J Hepatol. 1986;2:165-173. |
| 25. | Blackard JT, Sherman KE. Hepatitis C virus coinfection and superinfection. J Infect Dis. 2007;195:519-524. |
| 26. | Brown RJ, Juttla VS, Tarr AW, Finnis R, Irving WL, Hemsley S, Flower DR, Borrow P, Ball JK. Evolutionary dynamics of hepatitis C virus envelope genes during chronic infection. J Gen Virol. 2005;86:1931-1942. |
| 27. | Arenas JI, Gallegos-Orozco JF, Laskus T, Wilkinson J, Khatib A, Fasola C, Adair D, Radkowski M, Kibler KV, Nowicki M. Hepatitis C virus quasi-species dynamics predict progression of fibrosis after liver transplantation. J Infect Dis. 2004;189:2037-2046. |
| 28. | Qin H, Shire NJ, Keenan ED, Rouster SD, Eyster ME, Goedert JJ, Koziel MJ, Sherman KE. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood. 2005;105:533-541. |
| 29. | von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, McKeating JA. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132:667-678. |
| 30. | Torresi J, Stock OM, Fischer AE, Grollo L, Drummer H, Boo I, Zeng W, Earnest-Silveira L, Jackson DC. A self-adjuvanting multiepitope immunogen that induces a broadly cross-reactive antibody to hepatitis C virus. Hepatology. 2007;45:911-920. |
| 31. | Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata-Theimer ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106:7537-7541. |