Published online Aug 7, 2010. doi: 10.3748/wjg.v16.i29.3664
Revised: March 19, 2010
Accepted: March 26, 2010
Published online: August 7, 2010
AIM: To investigate and elucidate the molecular mechanism underlying varioliform gastritis for early detection, prevention and intervention of gastric cancer.
METHODS: A combination of two-dimensional gel electrophoresis and mass spectrometry was used to detect the differentially expressed proteins between varioliform gastritis and matched normal mucosa. The selected proteins were confirmed by Western blotting and reverse transcription polymerase chain reaction (RT-PCR) in additional samples and the function of some proteins in varioliform gastritis was analyzed by bio-method preliminarily.
RESULTS: We identified 21 differentially expressed proteins in varioliform gastritis, and compared them with matched normal mucosa. Eleven proteins were upregulated and ten downregulated in varioliform gastritis when compared with the same proteins in individual-matched normal gastric mucosa. These proteins are related to metabolism, oxidation, cytoskeleton, apoptosis, signal transduction and other aspects of cells. Two novel proteins, thioredoxin domain-containing protein 5 (TXNDC5) upregulated in varioliform gastritis, and neuropolypeptide h3 [phosphatidylethanolamine-binding protein 1 (PEBP1)] downregulated in varioliform gastritis, were further investigated. Their expressions were validated by Western blotting and RT-PCR in 12 cases of varioliform gastritis which was matched with normal mucosa. The expression level of PEBP1 in varioliform gastritis was significantly lower (P < 0.05) while that of TXNDC5 was significantly higher than that in matched normal gastric mucosa (P < 0.05).
CONCLUSION: There are some changes of protein expression in varioliform gastritis. Downregulation of PEBP1 and upregulation of TXNDC5 are involved in the development of varioliform gastritis.
-
Citation: Zhang L, Hou YH, Wu K, Zhai JS, Lin N. Proteomic analysis reveals molecular biological details in varioliform gastritis without
Helicobacter pylori infection. World J Gastroenterol 2010; 16(29): 3664-3673 - URL: https://www.wjgnet.com/1007-9327/full/v16/i29/3664.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i29.3664
Varioliform gastritis is currently recognized as a special kind of chronic gastritis characterized by nodules, thickened fugal folds and erosions. These features appear to be unusual and different from those seen in chronic gastritis. The diagnosis can be easily made by endoscopic examination. But the morbidity of varioliform gastritis has increased quickly recently in China. Very little is known about the etiopathogeny, clinical significance and evolution of this disease. The molecular biological researches on varioliform gastritis are very limited and no proteomics research on this disease has been found up to date. So the molecular mechanism of this disease is still unclear. The role of Helicobacter pylori (H. pylori) remains unknown. Although a close relationship between this gastritis and the bacteria was suggested to exist over the last few years, But no H. pylori infection was found in the gastric mucosa of some patients with varioliform gastritis. What is the reason?
Gastric cancer is the second most common malignancy in the world. Each year, about 798 000 people are diagnosed as having gastric cancer (9.9% of total cancer cases) and 628 000 people die from the disease (12.1% of cancer deaths)[1]. In eastern Asian countries including China, the morbidity and mortality of gastric cancer have ranked the first among all kinds of cancer and grown rapidly in the past two decades. Gastric carcinogenesis is not a well-known process, and the central paradigm for the initiation and development of gastric carcinoma is still not very clear.
In 1960, Munoz Monteavaro et al[2] reported varioliform gastritis with “in situ” carcinomatous transformation. It was reported a case of ampullary carcinoma accompanied with gastroenteropathy due to diffuse varioliform gastritis. Similarly, Cappell et al[3] reported adenomatous transformation in a patient with varioliform gastritis who had serial gastroscopies. This report also suggests a possible association between varioliform gastritis and gastric neoplasia. Several other groups have reported similar findings and performed a more comprehensive analysis of relationship between varioliform gastritis and gastric cancer[4-6].
The elevations could persist and appear as sessile polyps after the erosions heal and symptoms relieved after treatment. Adenomatous transformation was reported in some patients with varioliform gastritis. These reports suggested a possible association between varioliform gastritis and gastric neoplasia. Although this disease was concluded as a kind of precursor disease of gastric cancer at Sydney Conference, the mechanism of carcinogenesis from varioliform gastritis was unknown. Gastric cancer might be effectively controlled if this premalignant lesion-varioliform gastritis-is detected and treated before invasion occurs. Therefore, it is crucial to elucidate the molecular mechanism underlying varioliform gastritis. Some current mechanistic models focus almost on the localized lesion or H. pylori infection, with much less attention paid to pathologic changes occurring in the normal-appearing mucosa without H. pylori infection from which such lesions emerge.
The pattern of expressed proteins can reflect the information about the functional status and health of the tissue. Recently, the development of new methods for protein analysis has led to the emergence of a new field of clinical proteomics, in which these techniques are harnessed to identify functional molecular or biomarkers of cancer and other diseases[7], but there is hardly any study on the differential expressions of proteins in varioliform gastritis and normal-appearing mucosa.
In the present study, we used proteomic techniques to test the hypothesis that normal gastric mucosa from a patient with varioliform gastritis would exhibit different pattens of protein expression with the disordered mucosa from the same patient. By this approach, comparison of anatomically normal and disordered tissues against the same genetic background could be made.
Samples were taken from 17 patients with varioliform gastritis in the Second Affiliated Hospital of General Hospital of PLA (Table 1). These patients were examined by 13C urea breath test and the results were all negative. The results of autoantibody detection were also negative in these patients. The case of H. pylori infection and auto-immune disease was excluded. Normal gastric mucosa was defined as that 5cm adjacent to the elevations. All samples were obtained by biopsy in endoscopic examinations for these patients. Four pieces of elevatory tissues and normal mucosa were collected from each patient, respectively. One piece of the elevatory tissue underwent pathological diagnosis, and the others were saved for future studies. The patients were well informed in accordance with the disciplines of the Ethics Committee of Biomedicine, General Hospital of PLA, China.
| PatientNo. | Sex | Age (yr) | Lesion site | Histology |
| 1 | F | 77 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 2 | F | 54 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 3 | F | 59 | Gastric body and gastric antrum | Acute and chronic mucosal inflammation with Lymphocytic infiltration, H. pylori (-) |
| 4 | F | 43 | Gastric body and gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 5 | F | 62 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 6 | F | 68 | Gastric body and gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 7 | M | 44 | Gastric antrum | Acute and chronic mucosal inflammation with Lymphocytic infiltration, H. pylori (-) |
| 8 | M | 36 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 9 | M | 76 | Gastric antrum | Acute and chronic mucosal inflammation with Lymphocytic infiltration, H. pylori (-) |
| 10 | M | 67 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 11 | M | 55 | Gastric antrum and pylorus | Acute and chronic mucosal inflammation, H. pylori (-) |
| 12 | M | 45 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 13 | M | 72 | Gastric antrum | Acute and chronic mucosal inflammation with Lymphocytic infiltration, H. pylori (-) |
| 14 | M | 57 | Gastric antrum and pylorus | Acute and chronic mucosal inflammation, H. pylori (-) |
| 15 | M | 61 | Gastric antrum | Acute and chronic mucosal inflammation with Lymphocytic infiltration, H. pylori (-) |
| 16 | M | 51 | Gastric antrum | Acute and chronic mucosal inflammation, H. pylori (-) |
| 17 | M | 78 | Gastric antrum and pylorus | Acute and chronic mucosal inflammation, H. pylori (-) |
All samples were snap-frozen in liquid nitrogen and stored in a deep freezer (-80°C) until used. Tissues (80-150 mg) were crushed in liquid nitrogen and lysed in 1 mL of 7 mol/L urea, 2 mol/L thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonate (CHAPS), 65 mmol/L dithiothreitol (DTT), and 0.2% Bio-Lyte (pH 5-8, Bio-Rad, Hercules, CA) with sonication on ice. The lysates were centrifuged at 20 000 ×g for 1 h at 4°C. Supernatants were removed and concentrations were determined by the Bio-Rad AC DC protein assay kit (Bio-Rad). The protein samples were stored at -80°C. Before 2-DE was performed, the protein samples were purified using the Readyprep 2-D cleanup kit (Bio-Rad) according to the manufacturer’s instructions.
Detailed clinical and pathological data from the health care information center were reviewed. None of the patients had received treatment prior to endoscopic examination. Of the 17 patients, 11 were men, and six were women; the mean age was 51 years (range, 34-72 years, Table 1). No patient suffered from varioliform gastritis with other concurrent gastric diseases. All tissues of varioliform gastritis had definite histologic diagnoses: acute and chronic mucosal inflammation (n = 12), acute and chronic mucosal inflammation with lymphocytic infiltration (n = 5). None of them had H. pylori infection or low-to-moderate dysplasia.
Individual paired samples of normal gastric mucosa and varioliform gastritis were analyzed by 2-DE as described by Xing previously[8]. Briefly, linear gradient 24-cm (pH 5-8) ready strip (Bio-Rad) was rehydrated overnight at 16°C with 200 μg of protein samples in 500 μL of rehydration buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 65 mmol/L DTT, and 0.2% Bio-Lyte). Isoelectric focusing (IEF) was performed using PROTEAN IEF Cell (Bio-Rad). After IEF, the immobilized pH gradient strip was immediately equilibrated in equilibration buffer I [6 mol/L urea, 2% sodium dodecyl sulfate (SDS)], 0.375 mol/L Tris-HCl pH 8.8, 20% glycerol, and 2% DTT) for 15 min and then in equilibration buffer II (6 mol/L urea, 2% SDS, 0.375 mol/L Tris-HCl pH 8.8, 20% glycerol, and 2.5% iodoacetamide) for 15 min. SDS-polyacrylamide gel electrophoresis was carried out on 12% SDS-polyacrylamide gels (25 cm × 20.5 cm × 1.0 mm) using the PROTEAN Plus Dodeca Cell (Bio-Rad) at a constant voltage of 200 V at 20°C. After electrophoresis, the gels were stained using the Silver Stain Plus Kit (Bio-Rad). The above processes were performed in triplicate for each sample.
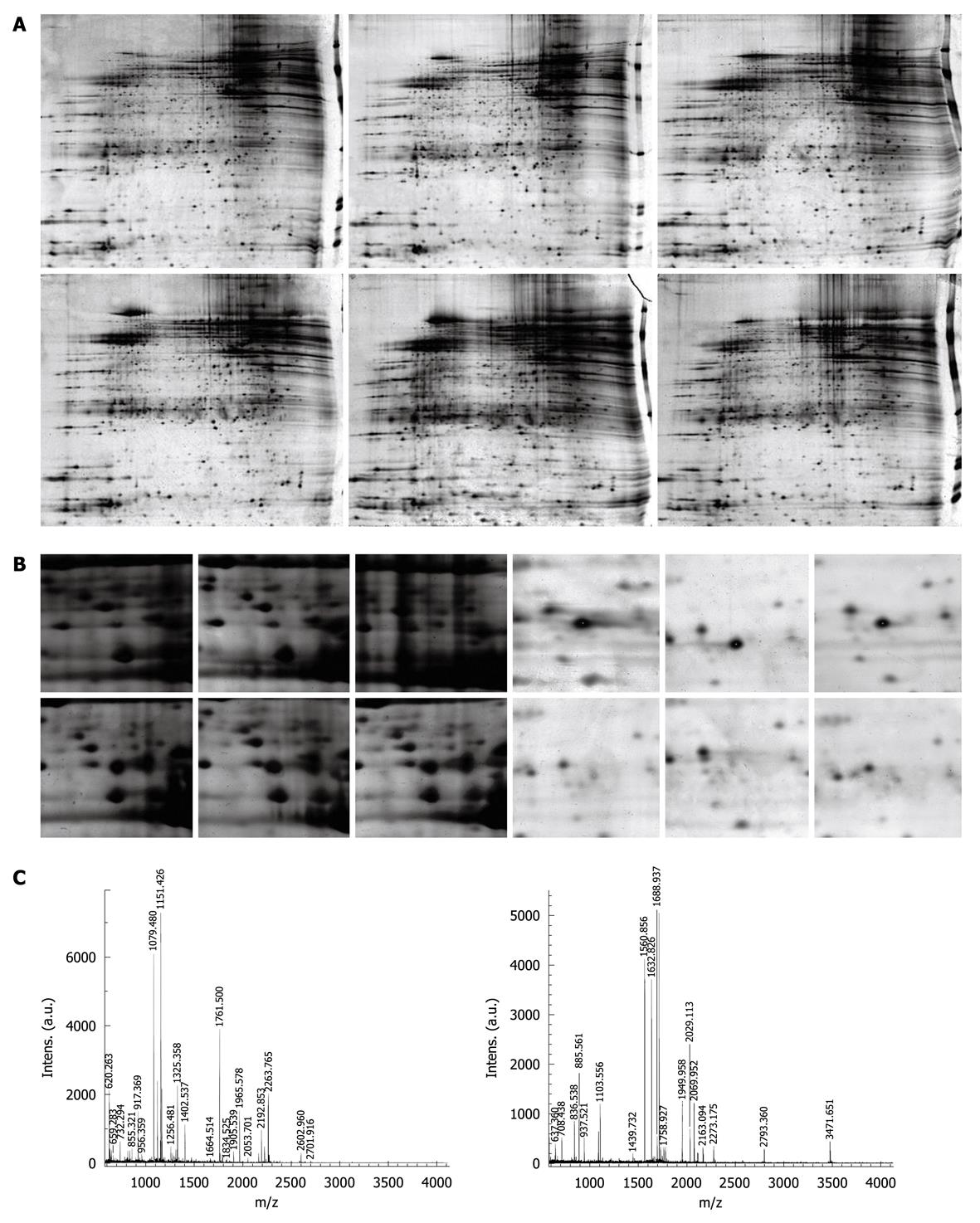
The silver-stained 2-DE gels were scanned on a GS-800 Calibrated Imaging Densitometer (Bio-Rad) at a resolution of 300 dots per inch. Intensities of protein spots were analyzed with Amersham Biosciences-Imagemaster v5.0. The differential protein spots were defined as those having a 5-fold higher or lower level of differential expression in at least 9 cases compared with the normal mucosa.
The 17 samples were used for spot cutting. Equal protein masses of each sample (normal gastric mucosa and varioliform gastritis tissue) were pooled, and 300 μg of the mixture was loaded for 2-DE. The differentially expressed protein spots were identified as described in the preceding text. These spots were excised from gels by Proteomeworks Spot Cutter (Bio-Rad), destained for 20 min in 30 mmol/L potassium ferricyanide/100 mmol/L sodium thiosulfate [1:1 (v/v)], and washed in Milli-Q water until the gels shrank and bleached. The gel pieces were incubated in 0.2 mol/L NH4HCO3 for 20 min and dried by lyophilization. Twenty microliters (20 μg/mL in concentration) trypsin (proteomics grade, Sigma, St. Louis, MO) was added into each gel piece, and incubated at 37°C overnight. The peptides were extracted three times with 50% acetonitrile and 0.1% trifluoroacetic acid and dried in a vacuum centrifuge.
The digests were analyzed using a Bruker Autoflex II TOF/TOF mass spectrometer with delayed extraction in which α-cyano-4-hydroxycinnamic acid was exploited as the matrix. The total 2-μL solution was applied onto a target disk and allowed to air-dry. Mass-to-charge ratios were measured in a reflector/delayed extraction mode with an accelerating voltage of 20 kV, a grid voltage of 63%-65%, positive polarity, and a delay time of 200 nanoseconds. Laser shots at 300 per spectrum were used to acquire the spectra from 800 to 4000 Daltons. Trypsin autolysis products were used for internal mass calibration. Database searching was performed using Mascot software (http://www.matrixscience.com). The search parameters were the nrNCBI database, human, 10-150 kDa, trypsin (1 missed enzymatic cleavage), and 100-ppm mass tolerance. The best match was the one with the highest score, and a significant match was typically a score of more than 70 (P < 0.05).
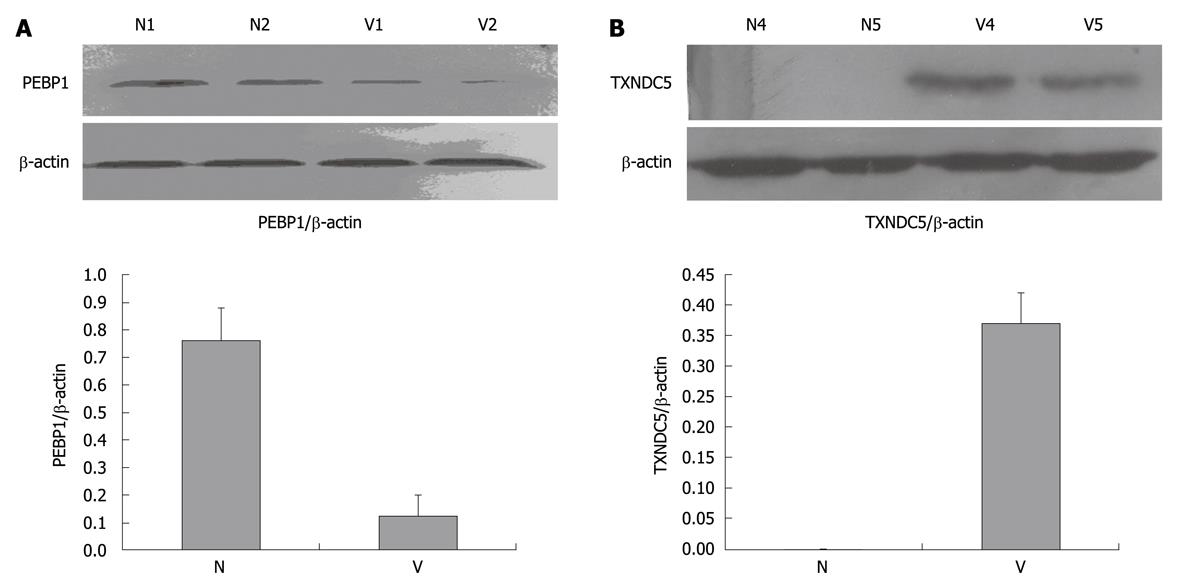
After the analysis of selected proteins, two differential proteins were confirmed by Western blotting analysis in additional samples for validating the 2-DE results. Western blotting analysis was performed in 12 cases of varioliform gastritis with individual-matched normal mucosa. Tissue samples were lysed as described above and protein extracts (50 μg) were separated on a 12% SDS-polyacrylamide gel. Proteins were transferred to a poly-vinylidene difluoride membrane (Bio-Rad). After blocking, the membranes were incubated with a rabbit monoclonal antibody of phosphatidylethanolamine-binding protein 1 (PEBP1) (dilution of 1:2000; Epitomics, California, MA) and polyclonal goat anti-thioredoxin domain-containing protein 5 (TXNDC5) antibody (dilution of 1:1000; Cell Signaling Technology, Danvers, MA). Subsequently, the membranes were incubated in horseradish peroxidase-anti-rabbit and horseradish peroxidase-anti-goat IgG (Abcam, Cambridge, UK), respectively. The specific proteins were visualized with chemiluminescent reagent (Pierce Biotechnology, Rockford, IL). As a control for equal protein loading, blots were restained with anti-actin antibody (dilution of 1:4000; Santa Cruz Biotechnology, Santa Cruz, CA). The band intensity was analyzed by PDQuest software v7.1. The relative expression level was calculated as the intensity ratio of PEBP1 or TXNDC5 to that of actin. The association between categorical data was analyzed using the SPSS11.0 software package.
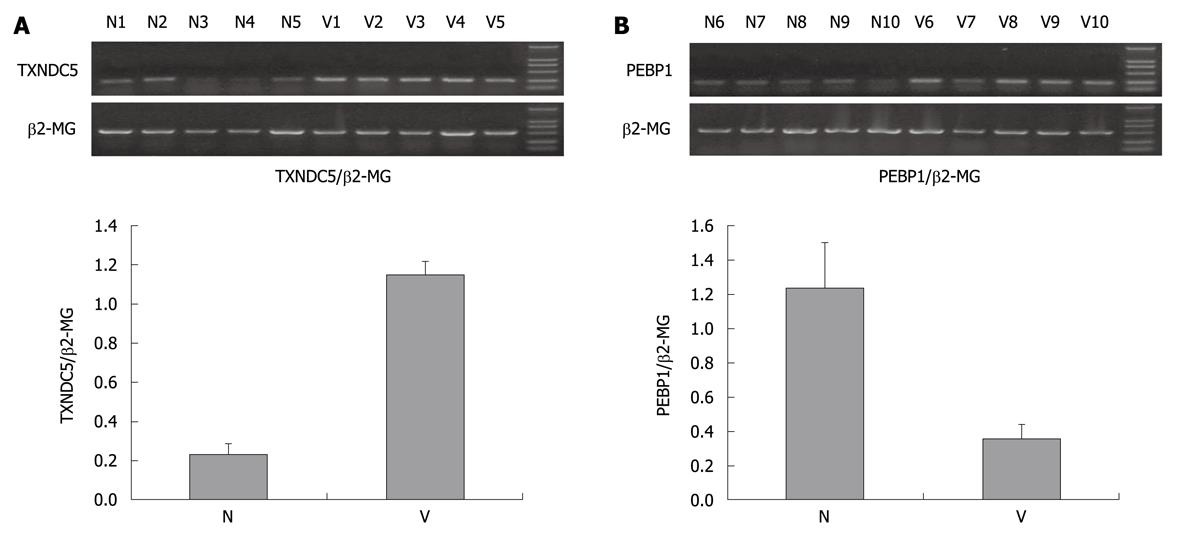
The total RNAs of additional samples were extracted by homogenization in Trizol (Invitrogen) for validating the 2-DE results. cDNA synthesis was performed in 20 μL reaction system of reverse transcription including 5 μg RNA. Amplification of TXNDC5, PEBP1 and β2-MG acting as internal control was carried out in DNA thermal cycler (Perkin Elmer) using equal cDNA as template. PCR products were separated by 1.5% agarose gel electrophoresis, scanned and analyzed with VDS ImageMaster system (Pharmacia).
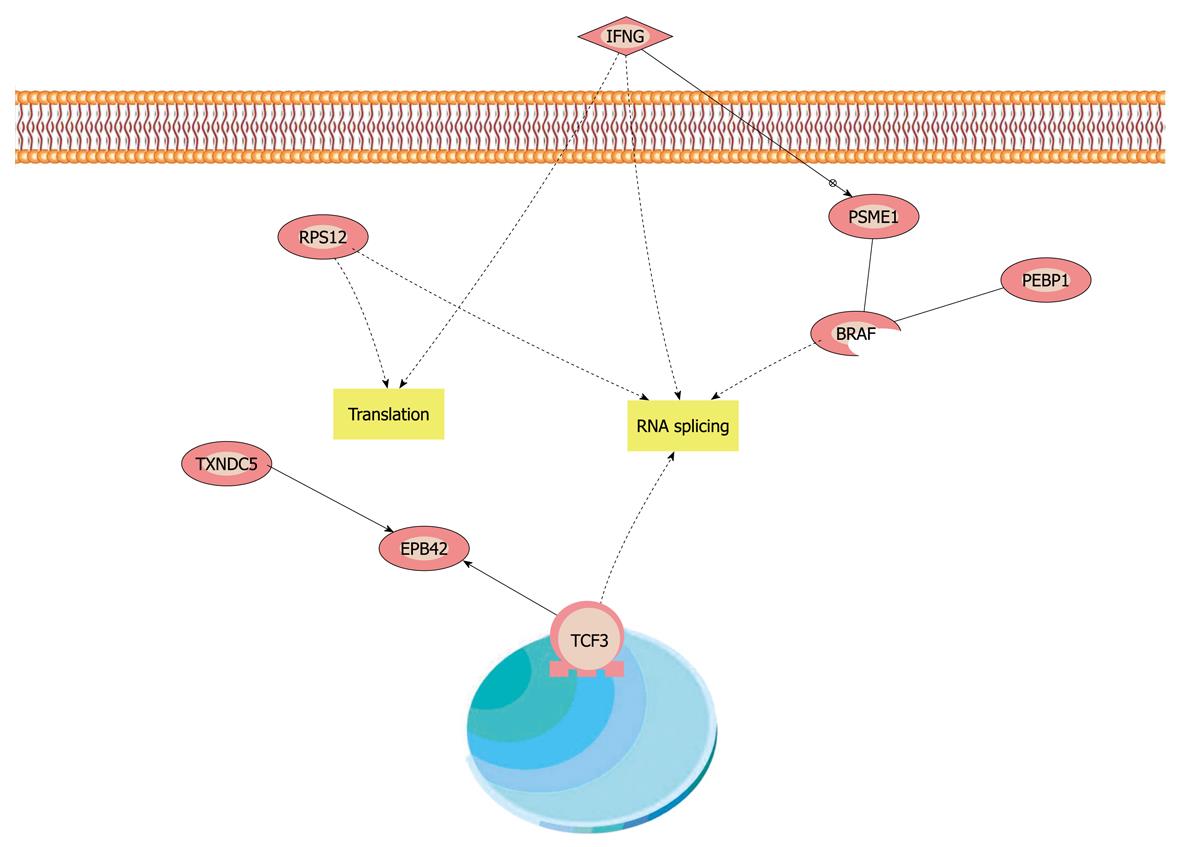
To understand the function of TXNDC5 and PEBP1 in varioliform gastritis, they were imported into Pathway Studio (demo), and a visualized interaction map was generated with information from Ensembl database, the Pfam protein family database, Prosite database, GNF GeneAtlas database and PDB database. Each node represents either a protein entity or a control mechanism of the interaction. We intended to find the key pathway including TXNDC5, PEBP1 and other proteins in our proteomics research by analyzing the protein interaction networks.
SPSS11.0 statistical software was used for the statistical analysis.
The gray values of the protein candidates were analyzed by the nonparametric Wilcoxon test. The intensity ratio of PEBP1 or TXNDC5 to that of internal control in Western blotting or reverse transcription polymerase chain reaction (RT-PCR) analysis was analyzed by one-factor analysis of variance.
The 2-DE protein patterns were studied in 17 patients with varioliform gastritis and individual-matched normal mucosa tissues. About 1800 proteins were detected in each gel. The proteins expressed in varioliform gastritis were compared with those in matched normal tissues. The differentially expressed candidates were the protein spots having a 5-fold higher or lower level of differential expression in at least 9 cases (Figure 1A and B). In this study, 21 significantly different candidate protein spots were found. They were also present in the 2-DE gel. Eleven proteins were upregulated and 10 downregulated in varioliform gastritis compared with the same proteins in individual-matched normal gastric mucosa. The quantities of all detected spots were analyzed by the nonparametric Wilcoxon test. These candidate spots were then analyzed by mass spectrometry (MS), and a total of 18 proteins (Figure 1C and Table 2) were identified. We failed to detect three protein spots. There might be several reasons, such as lower abundance, errors in the operation, lower reliability of the MS results, and characteristics of these proteins. More work will be done on the three protein spots in the future studies.
| ID No. | Protein name | Gene name | AccessionNo. | Mass (Da)/pI | Cover rate (mean, %) | Mascotscores | Intensity of candidateprotein spotsa (positive rate) | General function/comments |
| Up-regulated proteins | ||||||||
| 1 | Thioredoxin domain containing protein 5 precursor | TXNDC5 | Q8NBS9 | 47 629.2/5.3 | 25 | 228 | 36.5 ± 3.1, 2.7 ± 0.4 (100%, 44.4%) | Controlling the oxidative protein folding in endoplasmic reticulum |
| 2 | Proliferating cell nuclear antigen | PCNA | P12004 | 28 762.4/4.7 | 37 | 184 | 18.6 ± 4.2, 3.3 ± 0.9 (94.4%, 70.5%) | Cell growth and maintenance |
| 3 | 40S ribosomalprotein SA | RPSA | P08865 | 32 702.4/4.79 | 46 | 104 | 64.3 ± 11.1, 11.5 ± 2.8 (100%, 88.3%) | Originally known as laminin receptor precursor and p40 |
| 4 | Heat-shock protein β-1 | HSPB1 | P04792 | 22 768.5/5.9 | 33 | 73.8 ± 13.4, 12.7 ± 3.4 (100%, 100%) | A HSP27 isoform (pI5.68) | |
| 5 | Inorganic pyrophosphatase2 | PPA2 | Q9H2U2 | 35 472/5.9 | 69 | 245 | 24.1 ± 2.7, 3.6 ± 1.2 (94.4%, 58.9%) | Overexpressed in some cancer tissues |
| 6 | S100 calcium-binding protein A10 | S100A10 | P60903 | 11 195.5/6.8 | 62 | 148 | 43.0 ± 5.7, 5.9 ± 2.7 (88.9%, 41.2%) | It may function as a regulator of protein phosphorylation in the ANXA2 monomer |
| 7 | Nucleoside diphosphate kinase A | NME1 | P15531 | 17 137.7/5.8 | 49 | 193 | 59.4 ± 12.6, 13.2 ± 5.8 (82.4%, 70.6%) | It plays a major role in the synthesis of nucleoside triphosphates other than ATP |
| 8 | Proteasome activator complex subunit 1 | PSME1 | Q06323 | 38 966.2/7.6 | 53 | 138 | 83.8 ± 17.4, 10.5 ± 4.7 (100%, 41.2%) | Implicated in immuno-proteasome assembly and required for efficient antigen processing |
| 9 | Ubiquitin thiolesterase L3 | UCHL3 | P15374 | 26 337/4.7 | 42 | 174 | 62.4 ± 11.9, 13.7 ± 7.1 (88.9%, 29.4%) | Ubiquitin-protein hydrolase involved in the processing of both ubiquitin precursors and ubiquitinated proteins |
| 10 | S100 calcium-binding protein A6 | S100A6 | P06703 | 11 732.8/5.6 | 71 | 234 | 21.6 ± 5.3, 3.7 ± 1.1 (82.4%, 23.5%) | Preferentially expressed when quiescent fibroblasts are stimulated to proliferate |
| Down-regulated proteins | ||||||||
| 11 | Cell division cycle 2-like protein kinase 5 | CDC2L5 | Q14004 | 48 212.2/8.3 | 37 | 168 | 1.7 ± 0.5, 14.2 ± 2.6 (17.6%, 52.9%) | May be a controller of the mitotic cell cycle involved |
| 12 | BTG3 protein | BTG3 | Q14201 | 29 117.3/9.1 | 64 | 265 | 5.8 ± 2.3, 47.4 ± 6.1 (29.4%, 82.4%) | Overexpression impairs serum-induced cell cycle progression from the G0/G1 to S phase |
| 13 | Neuropolypeptide h3 | PEBP1 | P30086 | 31 270.6/5.7 | 58 | 220 | 2.9 ± 1.4, 58.6 ± 11.8 (17.6%, 100%) | Binds ATP, opioids and phosphatidylethanolamine |
| 14 | Heat-shock protein 17 kDa | HSPB3 | Q12988 | 16 966/5.7 | 39 | 105 | 16.8 ± 4.4, 89.5 ± 14.7 (100%, 100%) | Inhibitor of actin polymerization |
| 15 | Caspase-5 precursor | CASP5 | P51878 | 47 815/9.2 | 48 | 201 | 7.6 ± 2.6, 45.9 ± 11.7 (41.2%, 77.8%) | Mediator of apoptosis |
| 16 | Cytokeratin 20 | KRT20 | P35900 | 48 487/4.9 | 52 | 131 | 2.9 ± 0.4, 17.5 ± 3.6 (29.4%, 52.9%) | It plays a significant role in maintaining keratin filament organization in intestinal epithelia. When phosphorylated, it plays a role in the secretion of mucin in the small intestine |
| 17 | Eukaryotic translation initiation factor 3 subunit 2 | EIF3I | Q13347 | 23 354/4.9 | 44 | 173 | 8.9 ± 2.3, 46.3 ± 17.2 (35.3%, 100%) | Binds to the 40S ribosome and promotes the binding of methonyl |
| 18 | Ribosomal protein S12 | RPS12 | P25398 | 14 526.0/5.6 | 56 | 141 | 5.7 ± 1.4, 52.6 ± 10.3 (29.4%, 70.6%) | Belongs to the ribosomal protein S12e family |
The two novel candidate proteins, PEBP1 and TXNDC5, were studied further among the differentially expressed proteins. Their expression profiles in varioliform gastritis have not been reported previously. Western blotting analysis showed that TXNDC5 was upregulated significantly in varioliform gastritis but not in normal gastric mucosa (mean ± SD: 0.37 ± 0.05) (Figure 2B). Compared with that in normal mucosa (mean ± SD: 0.76 ± 0.12), PEBP1 was significantly downregulated in varioliform gastritis (mean ± SD: 0.18 ± 0.08) (P < 0.05, by Student’s test or the Friedman test) (Figure 2A).
Using semiquantitative RT-PCR, 476 bp fragment of TXNDC5, 451 bp fragment of PEBP1 and 876 bp control fragment of β2-MG were amplified (Figure 3). The mean ratios of the absorbency of PEBP1 band normalized to the control band were 0.35 ± 0.09 and 1.23 ± 0.27 in 12 cases of varioliform gastritis and normal mucosa. P value was lower than 0.05 when Student’s t test was used to compare the ratios of the two groups (Figure 3B). Those of TXNDC5 were 1.15 ± 0.07 and 0.23 ± 0.06 in 12 cases of varioliform gastritis and normal mucosa, respectively (P < 0.05) (Figure 3A). The results suggested that the difference of TXNDC5 and PEBP1 between varioliform gastritis and normal mucosa could be obvious at the mRNA level.
TXNDC5 and PEBP1 were imported into Pathway Studio (demo) to build an interaction network. The connectivity of TXNDC5 and PEBP1 was 40 and 145, respectively. The average connectivity of proteins identified was about 57. Our results showed that some members of mitogen-activated protein kinase (MAPK) family and some molecules involved in nuclear factor (NF)-κB and tumor necrosis factor were hot points with higher connectivity. From the mimical molecular network, we concluded that NF-κB, MAPK and interferon γ (IFN-γ) pathways were the cores of the whole network. The downstream-related cancer and other phenotypes were linked to the three pathways. The reaction of cell to IFN-γ could be the initiating agent of this molecular interaction network (Figure 4).
Because of limited knowledge on varioliform gastritis, the molecular events underlying this disease were still unknown, and the patients could be faced with more risk of gastric cancer. It was confirmed that the mucosal lesion of varioliform gastritis could develop into malignant tumor. Proteomic studies can help understand the early stages in the genesis of varioliform gastritis and has the potential to aid in the prevention and intervention for gastric cancer. In this study, we used the common approach of 2-DE coupled with MS to study the differentially expressed proteins in individual-matched cases of normal mucosa and lesion of varioliform gastritis and confirmed the differential expression of PEBP1 and TXNDC5 by Western blotting or RT-PCR.
A concept in cancer biology is that tumors arise and grow from some precancerous lesions as a result of the multiple changes of the genes or proteins which could influence the functions of cells via different molecular biological pathways. So it could be very important to find out these molecular changes and their functional pathways. The changes can be detected and analyzed in genomics and proteomics. A differential protein expression profile is a snapshot of the proteomics composition of a specific tissue at a specific time, which can be a key clue for further studies on the underlying mechanisms.
In this study, we identified 21 differentially expressed proteins in varioliform gastritis. However, none of these proteins (Table 2) had been reported in previous studies on this disease. We used a 5-fold cut-off according to the previous studies[9,10], and only found 21 differentially expressed proteins between varioliform gastritis and normal mucosa in the 2-DIGE study. We believe that some major molecular mechanisms underlying the disease should be implicated. There are also some methodological discrepancies in the process of our proteomic study, including the sample collection, the separation and identification of proteins and the analysis of results. Some low-abundance protein spots could not be displayed clearly, which should be further analyzed by a more advanced method.
PEBP1 expression was strong in normal mucosa, but significantly downregulated in varioliform gastritis. An alternative name of PEBP1 was Raf kinase inhibitory protein (RKIP) that belongs to the PEBP family. It is an inhibitor of the Raf/MEK/MAP kinase signaling cascade and is a suppressor of cancer metastasis[11]. Some researches[12,13] have confirmed that PEBP1 regulates activation of MAPK, NF-κB and G protein coupled receptors. As a modulator of key signaling pathways, PEBP1 affects various cellular processes, including cell differentiation, the cell cycle, apoptosis and cell migration. To date, emerging evidence[14-21] suggests that PEBP1 plays a crucial suppressing role in tumorigenesis and metastasis of prostate cancer, ovarian cancer, cervical cancer, colorectal cancer, liver cancer and breast cancer. It represents a novel effector of signal transduction pathways leading to apoptosis and a prognostic marker of the pathogenesis of human cancer cells and tumors. Chatterjee et al[22] have examined the expression patterns of PEBP1 and STAT3 in samples from 143 patients with gastric adenocarcinoma using tissue microarrays. Their results indicate the predictive and protective role of PEBP1 expression in gastric adenocarcinoma of the intestinal subtype. Downregulated expressions of PEBP1 could decrease patients’ survival. Collectively, these studies suggest that the PEBP1 or RKIP gene, as a potential tumor suppressor gene, is involved in gastric cancer initiation and progression, and expression of PEBP1 could be downregulated at initiation of tumorigenesis. Our results confirmed that PEBP1 expression was downregulated in varioliform gastritis compared with that of normal mucosa. We, therefore, postulate that this downregulation of PEBP1 might denote a step of the potential canceration.
TXNDC5 was significantly upregulated in varioliform gastritis although its expression remained in normal mucosa. TXNDC5 was first detected by 2-DE analysis of the luminal environment of the endoplasmic reticula of hepatic tissues in 2003[23]. As a novel PDI-like protein, TXNDC5 was highly expressed in endothelial cells. This tissue-specific expression is unusual among members of the PDI family. Sullivan et al[24] have confirmed that TXNDC5 could protect endothelial cells from stress-induced apoptosis. In contrast to PDI, which is essential for the survival of endothelial cells in the resting as well as the stressed state, TXNDC5 protects endothelial cells only under conditions of stress. They have found that loss of TXNDC5 results in reduced secretion of adrenomedullin and endothelin-1 together with a reduction in membrane-bound CD105, while TXNDC5 is essential for folding of CD105. The results of Edman’s and Freedman’s researches[25,26] suggested that TXNDC5 could play important roles in antioxidative injury, antianoxia-induced apoptosis, and promotion of proliferation in cells. Some recent studies showed that upregulation of TXNDC5 was found in tumors of the cervix, uterus, stomach and lung[24]. Nissom et al[27] found that a variant of the TXNDC5 gene was upregulated in poorly differentiated hepatocellular carcinoma (HCC) but unchanged in well-differentiated HCC. According to these reports, we think that TXNDC5 gene could be a tumor-enhancing gene, but the detailed biological roles of TXNDC5 in varioliform gastritis and gastric cancer remain to be elucidated. The upregulation of TXNDC5 in varioliform gastritis suggests that this disease could be related to gastric cancer, with higher risk than what was thought before.
The NF-κB and MAPK signaling pathways regulate growth in many tumors or inflammation, suggesting the cooperative role of these two pathways in the regulation of cell proliferation and apoptosis. H. pylori is known to be the cause of most gastric diseases, including both peptic ulcer disease and gastric cancer. Fox et al[28] think that the induction by H. pylori of cytokines and chemokines and growth-related genes is mediated by the MAPK and NF-κB signaling pathways, and Shibata et al[29] and Lee et al[30] have confirmed this conclusion. Kacar et al[31] and Chen et al[32] found that MAPK signaling pathway could be a causative factor in the alterations of the gastric mucosa infected by H. pylori and MAPK activation seems to be a significant and persistent event in the H. pylori-induced neoplastic transformation. IFN-γ acts through distinct cell surface receptors and induces transcription of an overlapping sets of genes. MHC class I genes are inducible by this interferon. IFN-γ is the gastric mucosal immunological reaction produced by T helper cells when gastric mucosa is infected by H. pylori[33-35]. It could induce the changes of TXNDC5 or PEBP1 and impact on the NF-κB and MAPK signaling pathways in our molecular interaction network of varioliform gastritis. We supposed that varioliform gastritis should be the results of a series of molecular interactions induced by IFN-γ or other molecules as an immunological reaction against microorganism infection according to previous reports and our research. But most of the previous studies focused on H. pylori, and the researches on other pathogens were very limited. So there could be some other bacteria or viruses which could induce an analogous immunological reaction against H. pylori in the gastric mucosa of varioliform gastritis patients without H. pylori infection.
In summary, our study showed a differential protein expression profile of varioliform gastritis compared with that of matched normal mucosa. The candidate proteins may confirm the previous conclusion that varioliform gastritis is one of the major precursor diseases of gastric cancer. The risk of potential canceration could be higher than what was thought previously, so effective treatment strategies should be studied and adopted for this disease in the future.
Varioliform gastritis is a chronic gastritis with a potential of developing into gastric cancer. To date, the etiopathogeny of the disease is unclear. So it is crucial to elucidate the molecular mechanism underlying the disease for preventing gastric cancer.
At present, the researches on varioliform gastritis focused mostly on endoscopic diagnosis and treatment or clinical feature. The reports on the molecular mechanism of the disease are very limited. As a new field of clinical proteomics is emerging, many new techniques have been developed to identify functional molecules or biomarkers of cancer and other diseases.
In this research, a differential protein expression profile of varioliform gastritis was indicated compared with that of matched normal mucosa. The important differential proteins and potential signal pathways have been provided for the future studies.
The results of this study provided some valuable clues for elucidating the molecular mechanism of varioliform gastritis and the relationship between the disease and gastric cancer. Some potential biomarkers were indicated for the early diagnosis of gastric cancer and therapeutic targets for this tumor.
Varioliform gastritis: a special kind of chronic gastritis characterized by nodules, thickened fugal folds and erosions. Although it is a very common gastritis, its features appear to be unusual and different from those seen in chronic gastritis.
The authors used proteomic techniques to identify differences in protein expression patterns in normal gastric mucosa vs mucosa characterized by varioliform gastritis. The study is well designed, represents a large amount of work, and will potentially be very helpful to further studies in the field of cancer research and treatment.
Peer reviewers: Dr. William R Parker, PhD, Assistant Professor, Department of Surgery, Duke University Medical Center, Box 2605, Durham, NC 27710, United States; Shashi Bala, PhD, Post doctoral Associate, Department of Medicine, LRB 270L, 364 Plantation street, UMass Medical School, Worcester, MA 01605, United States
S- Editor Tian L L- Editor Ma JY E- Editor Zheng XM
| 1. | Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33-64, 1. |
| 2. | Munoz Monteavaro C, Mendoza D, Palma E. [Varioliform gastritis with "in situ" carcinomatous transformation.]. An Fac Med Univ Repub Montev Urug. 1960;45:72-77. |
| 3. | Cappell MS, Green PH, Marboe C. Neoplasia in chronic erosive (varioliform) gastritis. Dig Dis Sci. 1988;33:1035-1039. |
| 4. | Gallina F, Benedetti-Valentini F. [Varioliform gastritis associated with gastric ulcer simulating a neoplasm.]. Riv Gastroenterol. 1963;15:85-94. |
| 5. | Vandenborre KM, Ghillebert GL, Rutgeerts LJ, Geboes KR, Rutgeerts PJ, Verbanck JJ, Tanghe WR. Hypertrophic lymphocytic gastritis with a gastric carcinoma. Eur J Gastroenterol Hepatol. 1998;10:797-801. |
| 6. | Mosnier JF, Flejou JF, Amouyal G, Gayet B, Molas G, Henin D, Potet F. Hypertrophic gastropathy with gastric adenocarcinoma: Menetrier's disease and lymphocytic gastritis? Gut. 1991;32:1565-1567. |
| 7. | Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267-275. |
| 8. | Xing X, Lai M, Gartner W, Xu E, Huang Q, Li H, Chen G. Identification of differentially expressed proteins in colorectal cancer by proteomics: down-regulation of secretagogin. Proteomics. 2006;6:2916-2923. |
| 9. | Stulík J, Hernychová L, Porkertová S, Knízek J, Macela A, Bures J, Jandik P, Langridge JI, Jungblut PR. Proteome study of colorectal carcinogenesis. Electrophoresis. 2001;22:3019-3025. |
| 10. | Roblick UJ, Hirschberg D, Habermann JK, Palmberg C, Becker S, Krüger S, Gustafsson M, Bruch HP, Franzén B, Ried T. Sequential proteome alterations during genesis and progression of colon cancer. Cell Mol Life Sci. 2004;61:1246-1255. |
| 11. | Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347-358. |
| 12. | Odabaei G, Chatterjee D, Jazirehi AR, Goodglick L, Yeung K, Bonavida B. Raf-1 kinase inhibitor protein: structure, function, regulation of cell signaling, and pivotal role in apoptosis. Adv Cancer Res. 2004;91:169-200. |
| 13. | Zeng L, Imamoto A, Rosner MR. Raf kinase inhibitory protein (RKIP): a physiological regulator and future therapeutic target. Expert Opin Ther Targets. 2008;12:1275-1287. |
| 14. | Woods Ignatoski KM, Grewal NK, Markwart SM, Vellaichamy A, Chinnaiyan AM, Yeung K, Ray ME, Keller ET. Loss of Raf kinase inhibitory protein induces radioresistance in prostate cancer. Int J Radiat Oncol Biol Phys. 2008;72:153-160. |
| 15. | Li HZ, Wang Y, Gao Y, Shao J, Zhao XL, Deng WM, Liu YX, Yang J, Yao Z. Effects of raf kinase inhibitor protein expression on metastasis and progression of human epithelial ovarian cancer. Mol Cancer Res. 2008;6:917-928. |
| 16. | Biewenga P, Buist MR, Moerland PD, Ver Loren van Themaat E, van Kampen AH, ten Kate FJ, Baas F. Gene expression in early stage cervical cancer. Gynecol Oncol. 2008;108:520-526. |
| 17. | Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of raf-1 kinase inhibitor protein expression is associated with tumor progression and metastasis in colorectal cancer. Am J Clin Pathol. 2007;127:820-827. |
| 18. | Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, García JJ, Scott L, Fyfe N, Murray GI. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol. 2006;24:5672-5679. |
| 19. | Lee HC, Tian B, Sedivy JM, Wands JR, Kim M. Loss of Raf kinase inhibitor protein promotes cell proliferation and migration of human hepatoma cells. Gastroenterology. 2006;131:1208-1217. |
| 20. | Schuierer MM, Bataille F, Weiss TS, Hellerbrand C, Bosserhoff AK. Raf kinase inhibitor protein is downregulated in hepatocellular carcinoma. Oncol Rep. 2006;16:451-456. |
| 21. | Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B, García JJ, Kolch W. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res. 2005;11:7392-7397. |
| 22. | Chatterjee D, Sabo E, Tavares R, Resnick MB. Inverse association between Raf Kinase Inhibitory Protein and signal transducers and activators of transcription 3 expression in gastric adenocarcinoma patients: implications for clinical outcome. Clin Cancer Res. 2008;14:2994-3001. |
| 23. | Clissold PM, Bicknell R. The thioredoxin-like fold: hidden domains in protein disulfide isomerases and other chaperone proteins. Bioessays. 2003;25:603-611. |
| 24. | Sullivan DC, Huminiecki L, Moore JW, Boyle JJ, Poulsom R, Creamer D, Barker J, Bicknell R. EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem. 2003;278:47079-47088. |
| 25. | Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267-270. |
| 26. | Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331-336. |
| 27. | Nissom PM, Lo SL, Lo JC, Ong PF, Lim JW, Ou K, Liang RC, Seow TK, Chung MC. Hcc-2, a novel mammalian ER thioredoxin that is differentially expressed in hepatocellular carcinoma. FEBS Lett. 2006;580:2216-2226. |
| 28. | Fox JG, Wang TC. Helicobacter pylori infection: pathogenesis. Curr Opin Gastroenterol. 2002;18:15-25. |
| 29. | Shibata W, Hirata Y, Ogura K, Omata M, Maeda S. [NF-kappaB and MAPK-signaling pathways contribute to the gene expression and host response induced by Helicobacter pylori infection]. Nippon Rinsho. 2005;63 Suppl 11:132-137. |
| 30. | Lee JS, Kim HS, Hahm KB, Sohn MW, Yoo M, Johnson JA, Surh YJ. Inhibitory effects of 7-carboxymethyloxy-3',4',5-trimethoxyflavone (DA-6034) on Helicobacter pylori-induced NF-kappa B activation and iNOS expression in AGS cells. Ann N Y Acad Sci. 2007;1095:527-535. |
| 31. | Kacar F, Meteoğlu I, Yasa H, Levi E. Helicobacter pylori-induced changes in the gastric mucosa are associated with mitogen-activated protein kinase (MAPK) activation. Appl Immunohistochem Mol Morphol. 2007;15:224-228. |
| 32. | Chen YC, Wang Y, Li JY, Xu WR, Zhang YL. H pylori stimulates proliferation of gastric cancer cells through activating mitogen-activated protein kinase cascade. World J Gastroenterol. 2006;12:5972-5977. |
| 33. | Cam S, Ertem D, Bahceciler N, Akkoc T, Barlan I, Pehlivanoglu E. The interaction between Helicobacter pylori and atopy: does inverse association really exist? Helicobacter. 2009;14:1-8. |
| 34. | Shimada M, Ando T, Peek RM, Watanabe O, Ishiguro K, Maeda O, Ishikawa D, Hasegawa M, Ina K, Ohmiya N. Helicobacter pylori infection upregulates interleukin-18 production from gastric epithelial cells. Eur J Gastroenterol Hepatol. 2008;20:1144-1150. |
| 35. | Vivas JR, Regnault B, Michel V, Bussière FI, Avé P, Huerre M, Labigne A, D' Elios MM, Touati E. Interferon gamma-signature transcript profiling and IL-23 upregulation in response to Helicobacter pylori infection. Int J Immunopathol Pharmacol. 2008;21:515-526. |