Published online Mar 14, 2010. doi: 10.3748/wjg.v16.i10.1267
Revised: March 27, 2009
Accepted: April 3, 2009
Published online: March 14, 2010
AIM: To establish the more feasible and sensitive assessment approach to the detection of adefovir (ADV) resistance-associated hepatitis B virus (HBV) quasispecies.
METHODS: Based on the characteristics of rtA181V/T and rtN236T mutations, a new approach based on real-time fluorescent quantitative polymerase chain reaction (RT-PCR) was established for the detection of ADV-resistant HBV quasispecies, total HBV DNA, rtA181 and rtN236 mutations in blood samples from 32 chronic hepatitis B (CHB) patients with unsatisfactory curative effect on ADV and compared with routine HBV DNA sequencing.
RESULTS: Both the sensitivity and specificity of this new detection approach to ADV-resistant HBV quasispecies were 100%, which were much higher than those of direct HBV DNA sequencing. The approach was able to detect 0.1% of mutated strains in a total plasmid population. Among the 32 clinical patients, single rtA181 and rtN236T mutation and double rtA181T and rtN236T mutations were detected in 20 and 8, respectively, while ADV-resistant mutations in 6 (including, rtA181V/T mutation alone in 5 patients) and no associated mutations in 26.
CONCLUSION: This new approach is more feasible and efficient to detect ADV-resistant mutants of HBV and ADV-resistant mutations before and during ADV treatment with a specificity of 100% and a sensitivity of 100%.
- Citation: Zhao WF, Shao YL, Chen LY, Wu JH, Zhu YL, Gan JH, Xiong H. Establishment of a new quantitative detection approach to adefovir-resistant HBV and its clinical application. World J Gastroenterol 2010; 16(10): 1267-1273
- URL: https://www.wjgnet.com/1007-9327/full/v16/i10/1267.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i10.1267
Chronic hepatitis B virus (HBV) infection affects approximately 400 million people worldwide and is still an important cause of morbidity and mortality, as well as a source of potential new infections[1]. Chronic HBV carriers may develop chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) with HBV as the most frequent cause[2]. Since chronic hepatitis B (CHB) patients require a prolonged if not a lifelong therapy, data about its long-term safety and effect are indispensable for the assessment of its risk and benefit. Treatment of CHB has been improved due to available nucleoside/tide analogues, such as lamivudine (LAM), adefovir dipivoxil (ADV), entecavir (ETV), and telbivudine (LdT), which act as inhibitors of HBV reverse transcriptase and decrease viral load in most cases[3]. ADV has become a treatment option for HBV infection due to its effect on lamivudine-resistant mutations occurring upon prolonged treatment. However, viral resistance to ADV develops and increases over time. ADV-resistance is mainly associated with rtN236T and rtA181V/T mutations within the D and B functional domains of HBV polymerase[4]. Many methods have been developed to detect ADV-resistant mutations, such as direct polymerase chain reaction (PCR) sequencing, INNO-LiPA, restriction fragment length polymorphism (PCR-RFLP), matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI TOF MS)[4-9], with their respective advantages and disadvantages. Sequencing remains the best approach to the identification of new mutations. However, it cannot detect rtN236T and rtA181V/T mutations in less than 25% of a total viral population and is not appropriate for large-scale use in large cohort studies or clinical laboratories because of its labor-intensive and time-consuming manipulations[5]. INNO-LiPA and MALDI TOF MS are capable of detecting variants, but more strict experiment conditions and equipments are required. PCR-RFLP[8] could only detect mutations in a high proportion and is also labor-intensive and time-consuming. This study described a feasible and sensitive approach to quantitative detection of ADV-resistant HBV quasispecies by real-time fluorescent quantitative PCR.
Thirty-two patients (29 males and 3 females, mean age 35 years, range 20-63 years) who were diagnosed as CHB from March 2003 to October 2007 in our hospital following the guidelines of prevention and treatment of chronic hepatitis B (2005)[10] were enrolled in this study. ADV was administered with oral dipivoxil (10 mg) for 58 wk (range: 42-82 wk), during which viral breakthrough, viral rebound, partial or inadequate virus response occurred[11]. Exclusion criteria included a coexisting severe illness, organ or bone marrow transplantation, recent treatment with systemic corticosteroids, immunosuppressants or chemotherapeutic agents, liver disease not due to hepatitis B, and seropositivity for human immunodeficiency virus (HIV) or hepatitis C or D virus. Informed consent was obtained from each patient in accordance with the Declaration of Helsinki. The study was approved by the institutional review board of Soochow University. The average load of HBV DNA at baseline before ADV therapy was 6.30 ± 0.23 log10 IU/mL, and dropped to 5.61 ± 0.17 log10 IU/mL after treatment with ADV for 82 wk.
HBV DNA was extracted from 100 μL of sera as previously described[6]. Extracted DNA pellets were resuspended in 30 μL of sterile distilled water and stored at -20°C until use. HBV DNA fragments containing the RT domain were amplified by PCR using special primers (RTS: 5'AAAATCCTCACAATACCAC3', RTSA: 5'GACATACTTTCCAATCAATAG3') as previously described[6]. The PCR products were cloned into the pUC-18 vector (TAKARA) following its manufacturer’s instructions. Following transformation into DH5α competent cells (Tiangen, Shanghai), each wild strain plasmid, rtA181V/T and rtN236T variant plasmid of types B and C were picked out respectively according to the sequencing results, and standardized with a quantitative test kit for HBV DNA. After serial dilutions, quantitative reference substances for related rt181 and rt236 variant mutations were prepared.
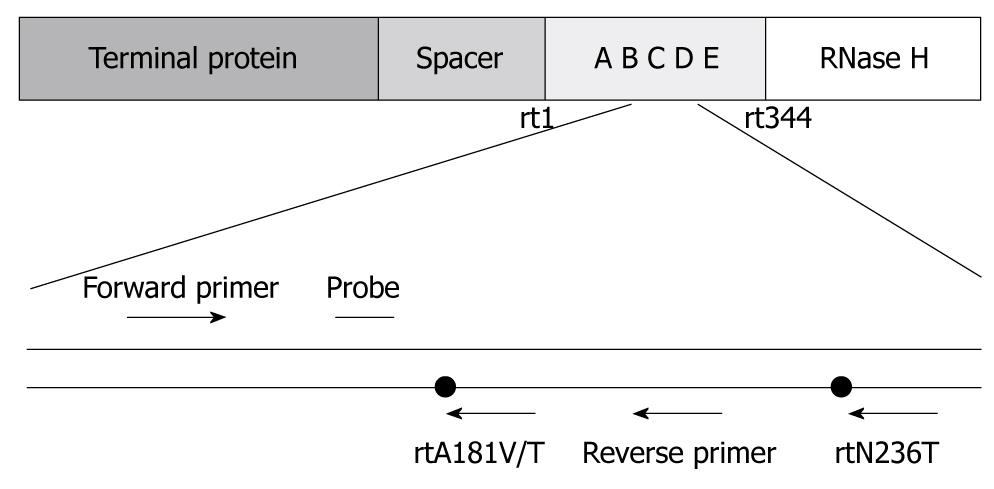
Based on the characteristics of rtA181V/T and rtN236T mutations, specific primers and Taqman probes were designed (Figure 1). Mutations of rtA181 and rtN236 and total HBV DNA isolated from the same serum specimens were detected by real-time fluorescent quantitative PCR as previously described[7].
Primers for PCR were designed to target highly conservative sequences of HBV gene in oligonucleotides: P1, 5'CCTGCTGGTGGCTCCAGTTCAGGAACAG3' (nt56-83), P2, 5'AAGCCCCAACCAGTGGGGGTTGCGTCA3' (nt1188-1214). The amplified fragments contained the polymerase RT domain and overlapped the S gene. Viral DNA was amplified with a HBV sequencing kit (Shenyou, Shanghai, China). Positive PCR products identified by agarose gel electrophoresis were directly sequenced with an ABI PRISM 3730 genetic analyzer.
Mutations of rtA181 or rtN236 were assayed with the ratio of total HBV DNA to rtA181 or rtN236 mutation-associated DNA. The assessment criteria used were as follows: (1) If the ratio (a/b or a/c) of total DNA to rtA181 or rtN236 mutation-associated DNA was between 1 and 10, it was defined as complete variants or the proportion of mutations was more than 10% in the pool of quasispecies; (2) If the ratio (a/b or a/c) of total DNA to rtA181 or rtN236 mutation-associated DNA was between 10 and 200, it was defined as the proportion of variants was lower than 10% in the pool of quasispecies; and (3) If the ratio (a/b or a/c) of total DNA to rtA181 or rtN236 mutation-associated DNA was more than 200, it was defined as complete wild strains or the proportion of variants was below the limit of identification.
All data were analyzed using the SAS V8.1 statistical package version. Analysis of one way variance (ANOVA) and t-test were used to compare the average value. P < 0.05 was considered statistically significant
Each plasmid listed in Table 1 with a concentration of 6 × 106 IU/mL was picked out for real-time fluorescent quantitative PCR. The rtN236T and rtA181were found to be wild type plasmids at positions 181 and 236, respectively (Table 1), indicating that the new approach has a sensitivity of 100% and a specificity of 100%.
| Plasmid | HBV DNA reagent | rtA181 reagent | rtN236 reagent |
| Wild strain plasmid of Type B | 7.27E+06 | - | - |
| Wild strain plasmid of Type C | 7.23E+06 | - | - |
| rtA181V variant plasmid | 8.78E+06 | 1.88E+06 | - |
| rtA181T variant plasmid | 1.14E+07 | 2.26E+06 | - |
| rtN236T variant plasmid of Type B | 1.14E+07 | - | 1.25E+06 |
| rtN236T variant plasmid of Type C | 1.12E+07 | - | 1.31E+06 |
| Mixed variants plasmid | 5.92E+06 | 2.00E+06 | 1.18E+06 |
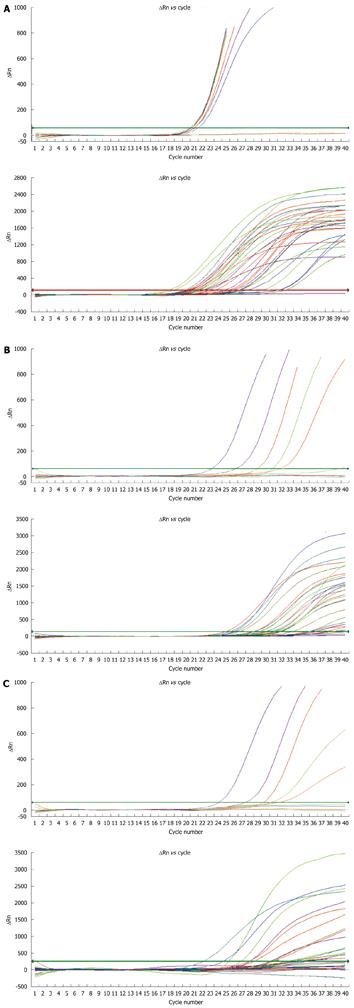
Variant and wild type plasmids with a concentration of 6 × 106 IU/mL were picked out and diluted at 0/1, 1/1, 1/10, 1/50, 1/200, 1/1000, and 1/10 000, respectively. The results of fluorescent quantitative PCR are shown in Figure 2A and Table 2. The Ct value for the wild type plasmids was different.
| Variant: wild strain | HBV DNA reagent | rtA181 reagent | rtN236 reagent | a/b | a/c |
| 0:1 | 6.06E+06 | - | - | ||
| 1:1 | 2.94E+06 | 4.80E+06 | 5.10E+06 | 0.61 | 0.58 |
| 1:10 | 3.98E+06 | 486 276.3 | 337 566.8 | 8.18 | 11.79 |
| 1:50 | 4.28E+06 | 83 242.16 | 85 867.71 | 51.42 | 49.84 |
| 1:200 | 5.63E+06 | 23 130.8 | 22 000 | 243.4 | 255.91 |
| 1:1000 | 5.76E+06 | 7880 | 6223.36 | 730.96 | 925.55 |
| 1:10 000 | 6.62E+06 | - | - |
Total HBV DNA, and rtA181 or rtN236 mutations in 8 serum samples from the patients who did not receive ADV treatment and had no associated mutations detected by sequencing were detected by fluorescent quantitative PCR. The ratio(a/b or a/c) of total HBV DNA to rtA181 or rtN236 mutation-associated DNA was lower than 1000 when the variant plasmids were mixed with wild type plasmids at a dilution of 1:1000 and higher than 5000 in the patients who did not receive ADV therapy (Table 3).
| Serum samples | HBV DNA reagent | rtA181 reagent | rtN236 reagent | a/b | a/c |
| Sample 1 | 2.45E+07 | 541.19 | - | 45 270.61 | |
| Sample 2 | 6.79E+06 | - | - | ||
| Sample 3 | 1.09E+07 | 935.07 | - | 11 656.88 | |
| Sample 4 | 7.48E+06 | - | - | ||
| Sample 5 | 3.24E+07 | 4285.91 | 185.27 | 7559.65 | 174 879.91 |
| Sample 6 | 3.02E+07 | 1652.39 | - | 18 276.56 | |
| Sample 7 | 4.22E+06 | - | - | ||
| Sample 8 | 1.64E+07 | 2871.36 | - | 5711.58 |
The ratio of total HBV DNA to rtA181 and rtN236 mutation-associated DNA (a/b or a/c) in mutations of variant and wild- type plasmids was less than 1000 at a dilution of 1:1000, while the ratio of total HBV DNA to ADV-resistant mutations was higher than 5000 in the naïve patients. Among the 32 patients, mutations of rtA181 and rtN236T were detected in 20 and 8, respectively. Based on the current assessment criteria (the ratio of a/b cut-off was 1, 10 and 200, respectively), the 32 patients were divided into 3 groups with the a/b ratio = 1 to 10 in 6, = 10 to 200 in 7 and ≥ 200 in 12 patients, respectively, and the a/c ratio = 1 to 10 in 1, = 10 to 200 in 7 and ≥ 200 in 24 patients, respectively. The rtA181 mutation level in HBV DNA was 6.22 ± 0.55 log10 IU/mL, 4.78 ± 0.21 log10 IU/mL, and 3.14 ± 0.33 log10 IU/mL, respectively, in these three groups (P < 0.0001). The rtN236 mutation level in HBV DNA was 5.31 ± 0.25 log10 IU/mL and 2.49 ± 0.19 log10 IU/mL, respectively, in patients with their a/c ratio = 1 to 200 or ≥ 200 (P < 0.0001) (Figure 2B and Table 4).
| a/b | n | rtA181 mutation (log10 IU/mL) | P1 | a/c | n | rtN236 mutation (log10 IU/mL) | P2 |
| 1-10 | 6 | 6.22 ± 0.55 | < 0.0001 | 1-200 | 8 | 5.31 ± 0.25 | < 0.0001 |
| 10-200 | 14 | 4.78 ± 0.21 | |||||
| ≥ 200 | 12 | 3.14 ± 0.33 | ≥ 200 | 24 | 2.49 ± 0.19 |
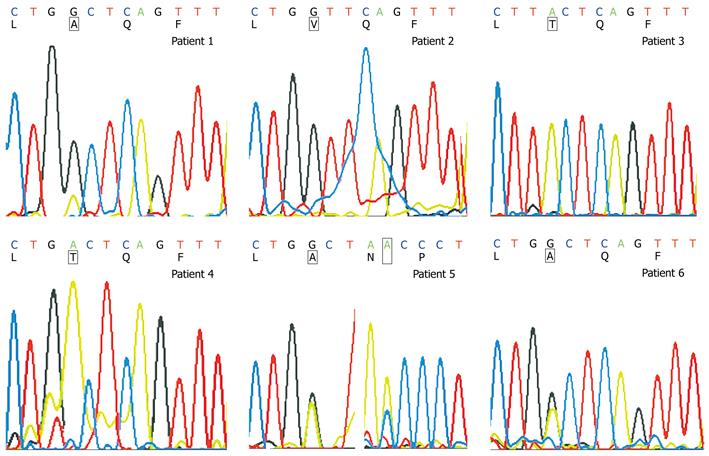
Of the 6 patients with ADV-resistant mutations, rtA181T and rtN236T mutations and single rtA181V/T mutation were observed in patients 1 and 5, respectively. Among the 26 patients with no associated mutations, rtA181 variants and wild type strains were found in patients 1, 5 and 6 (Figure 3) while only rtA181variants were detected in patients 2, 3 and 4 (Table 5).
Dipivoxil, a nucleotide analogue, has become a treatment option for HBV infection due to its efficacy on lamivudine-resistant mutations occurring upon prolonged treatment. However, resistance to ADV develops and increases over time. ADV therapy for naive patients is associated with delayed and less frequent drug-resistant mutations. The reported cumulative incidence of mutations in hepatitis B e antigen (HBeAg)-negative and positive chronic hepatitis B patients is 29% and 20%, respectively after 5 years of ADV treatment[12,14]. LAM-resistant patients treated with ADV monotherapy are at a higher risk of developing ADV-resistant mutations than nucleoside-naive patients[7,13]. In accordance with the current guidelines, ADV should be used in combination with other drugs in treatment of LAM-resistant patients instead of ADV monotherapy. It has been shown that ADV-resistant mutants occur infrequently in patients on combined therapy[15]. An open-label study in HBeAg-negative LAM-resistant patients demonstrated that combined therapy does not result in development of resistance to ADV over a period of 3 years[16]. It has been recently reported that the rate of ADV resistance is 1% and 4% 1 and 3 yeas after ADV + LAM therapy[17]. Two mutations in the HBV polymerase gene, alanine to valine or threonine (rtA181V/T) and asparagine to threonine substitution (rtN236T) have been described to confer resistance to ADV[4], suggesting that genotypic resistance monitoring is important to anticipate and confirm the observed phenotypic resistance and new treatment modalities can be selected. The incidence of genotypic resistance may vary with the sensitivity of different methods used to detect resistant mutations. Thus, methodological options for detecting HBV mutations are an increasingly important issue in clinical management of CHB. Several other methods have been used to detect ADV-resistant HBV. However, the disadvantages of each method restrict its wide-range use. So developing a more sensitive, specific and easy approach is always the major concern for researchers. Malmström et al[18] developed a method based on real-time polymerase chain reaction with TaqMan chemistry which can target wild type and mutant viral strains at rtL180 and rtM204, later, Lupo et al[19] described a selective real-time PCR (sPCR) for the quantitative detection of rtM204I/V and rtN236T mutations associated with resistance to LAM and ADV, with a lowest limit of 0.1% of strains carrying rtM204V/I or rtN236T mutations for a total of 105 copies of plasmid. In this study, primers and probes for quantitative amplification of HBV DNA, rtA181 and rtN236 mutations were designed, and the results showed that they could detect more than 100 IU/mL of variants. Mutants could be obtained by calculating the ratio of total HBV DNA to the variants. To assess the feasibility of this approach, HBV DNA, rtA181 and rtN236 mutations were detected in 32 patients with unsatisfactory response to adefovir dipivoxil by direct sequencing. The results showed that this new approach could detect rtA181 and rtN236 mutations as well as some ADV mutations not identified by direct sequencing, thus supporting the new approach with a high concordance and a higher sensitivity. ADV-resistant HBV mutations can be pre-monitored, which can provide valuable reference for shifting the treatment in order to prevent hepatitis deterioration and decompensation. Furthermore, real-time fluorescent quantitative PCR can be widely used.
Meanwhile, there is a large room for the improvement of this new approach. The major improvement in RT-PCR is to design the optimal primers for each mutant. Moreover, due to the variability of genotypes, a number of primers may be required for the detection of a single nucleotide polymorphism. In this study, the primers were restricted to the most frequent codons associated with rtA181 and rtN236T mutations. However, further study is needed to verify the other mutations.
In conclusion, the new approach based on real-time fluorescent quantitative PCR is more feasible than direct DNA sequencing. It is more sensitive and specific for the detection of wild-type and variant viruses as well as ADV-resistant mutations. We believe that this easy-to-use approach will play an important role in detection of ADV-resistant mutations and in optimization of ADV therapy.
With the availability of more potent nucleotide/nucleoside analogues, early detection of drug-resistant mutants of hepatitis B virus (HBV) is important for the treatment of chronic hepatitis B (CHB). The incidence of genotypic resistance may vary with the sensitivity of methods used to detect resistant mutations. Thus, methodological options for detecting HBV mutations are an increasingly important issue in clinical management of CHB.
The study described a practical and accurate method for quantitative detection of adefovir (ADV)-resistant HBV quasispecies using real-time fluorescent quantitative polymerase chain reaction (PCR). The results demonstrated that this method could detect and monitor ADV resistance mutations in patients chronically infected with HBV and is more sensitive than sequencing.
ADV-resistant HBV quasispecies can be detected by real-time fluorescent quantitative PCR, which should be extensively popularized in clinical practice.
The method described in this paper is specific, sensitive and accurate for the detection of ADV-resistant mutants of HBV in ADV-treated patients, thus adding a new tool for monitoring ADV-resistant mutations before and during ADV therapy.
Peer reviewer: Juan-Ramón Larrubia, PhD, Gastroenterology Unit and Liver Research Unit., Guadalajara University Hospital, Donante de Sangre s/n, 19002 Guadalajara, Spain
S- Editor Cheng JX L- Editor Wang XL E- Editor Zheng XM
| 1. | Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335-352. |
| 2. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. |
| 3. | Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254-265. |
| 4. | Delaney WE 4th. Progress in the treatment of chronic hepatitis B: long-term experience with adefovir dipivoxil. J Antimicrob Chemother. 2007;59:827-832. |
| 5. | Niesters HG, Pas S, de Man RA. Detection of hepatitis B virus genotypes and mutants: current status. J Clin Virol. 2005;34 Suppl 1:S4-S8. |
| 6. | Osiowy C, Villeneuve JP, Heathcote EJ, Giles E, Borlang J. Detection of rtN236T and rtA181V/T mutations associated with resistance to adefovir dipivoxil in samples from patients with chronic hepatitis B virus infection by the INNO-LiPA HBV DR line probe assay (version 2). J Clin Microbiol. 2006;44:1994-1997. |
| 7. | Yeon JE, Yoo W, Hong SP, Chang YJ, Yu SK, Kim JH, Seo YS, Chung HJ, Moon MS, Kim SO. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006;55:1488-1495. |
| 8. | Chen JJ, Ma SW, Wang ZH, Sun J, Hou JL. [Kinetics of HBV mutants conferring adefovir resistance (rtn236t) and a method to detect them rapidly]. Zhonghua Ganzangbing Zazhi. 2008;16:33-37. |
| 9. | Hong SP, Kim NK, Hwang SG, Chung HJ, Kim S, Han JH, Kim HT, Rim KS, Kang MS, Yoo W. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J Hepatol. 2004;40:837-844. |
| 10. | Chinese Society of Hepatology, Chinese Society of Infectious Diseases. The guideline of prevention and treatment for chronic hepatitis B. Zhonghua Neike Zazhi. 2006;45:162-170. |
| 11. | Keeffe EB, Zeuzem S, Koff RS, Dieterich DT, Esteban-Mur R, Gane EJ, Jacobson IM, Lim SG, Naoumov N, Marcellin P. Report of an international workshop: Roadmap for management of patients receiving oral therapy for chronic hepatitis B. Clin Gastroenterol Hepatol. 2007;5:890-897. |
| 12. | Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, Chang TT, Kitis G, Rizzetto M, Marcellin P, Lim SG, Goodman Z, Ma J. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology. 2006;131:1743-1751. |
| 13. | Lee YS, Suh DJ, Lim YS, Jung SW, Kim KM, Lee HC, Chung YH, Lee YS, Yoo W, Kim SO. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology. 2006;43:1385-1391. |
| 14. | Marcellin P, Chang TT, Lim SG, Sievert W, Tong M, Arterburn S, Borroto-Esoda K, Frederick D, Rousseau F. Long-term efficacy and safety of adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2008;48:750-758. |
| 15. | Peters MG, Hann Hw H, Martin P, Heathcote EJ, Buggisch P, Rubin R, Bourliere M, Kowdley K, Trepo C, Gray Df D. Adefovir dipivoxil alone or in combination with lamivudine in patients with lamivudine-resistant chronic hepatitis B. Gastroenterology. 2004;126:91-101. |
| 16. | Rapti I, Dimou E, Mitsoula P, Hadziyannis SJ. Adding-on versus switching-to adefovir therapy in lamivudine-resistant HBeAg-negative chronic hepatitis B. Hepatology. 2007;45:307-313. |
| 17. | Lampertico P, Viganò M, Manenti E, Iavarone M, Sablon E, Colombo M. Low resistance to adefovir combined with lamivudine: a 3-year study of 145 lamivudine-resistant hepatitis B patients. Gastroenterology. 2007;133:1445-1451. |
| 18. | Malmström S, Hannoun C, Lindh M. Mutation analysis of lamivudine resistant hepatitis B virus strains by TaqMan PCR. J Virol Methods. 2007;143:147-152. |
| 19. | Lupo J, Larrat S, Hilleret MN, Germi R, Boyer V, Nicod S, Barguès G, Leroy V, Seigneurin JM, Zarski JP. Assessment of selective real-time PCR for quantitation of lamivudine and adefovir hepatitis B virus-resistant strains and comparison with direct sequencing and line probe assays. J Virol Methods. 2009;156:52-58. |