Published online Sep 7, 2009. doi: 10.3748/wjg.15.4183
Revised: August 2, 2009
Accepted: August 9, 2009
Published online: September 7, 2009
AIM: To investigate the expression and clinical significance of S100A2 mRNA and protein, p63 protein in esophageal squamous cell carcinoma (ESCC) and their roles in carcinogenesis and progression of esophageal carcinoma (EC).
METHODS: Immunohistochemical staining (S-P method) for S100A2 and p63 protein were performed in 40 samples of ESCC and 40 samples of normal esophageal mucosa. In situ hybridization (ISH) was used to detect the expression of S100A2 mRNA.
RESULTS: Expression of S100A2 mRNA in ESCC was positive in 77.5% of samples, which was lower than that in normal mucosa (100%) by ISH (P = 0.002). The expression level of S100A2 mRNA was closely related to differentiation and and node-metastasis (P = 0.012, P = 0.008). Expression of S100A2 protein was positive in 72.5% of ESCC samples and expression of p63 protein was positive in 37.5% of ESCC samples, and was lower than that in normal mucosa (100%) (P = 0.000). The expression of S100A2 protein was correlated with the differentiation and node-metastasis (P = 0.007, P = 0.001), but no relationship was observed between the expression of p63 protein and clinical pathological manifestations. S100A2 protein was positively correlated with the expression of S100A2 mRNA, and negatively associated with the expression of p63 protein (P = 0.000, P = 0.002).
CONCLUSION: S100A2 and p63 protein both play important roles in the carcinogenesis of ESCC. An investigation into the combined expression of S100A2 and p63 may be helpful in early diagnosis and in evaluating the prognosis of ESCC.
- Citation: Cao LY, Yin Y, Li H, Jiang Y, Zhang HF. Expression and clinical significance of S100A2 and p63 in esophageal carcinoma. World J Gastroenterol 2009; 15(33): 4183-4188
- URL: https://www.wjgnet.com/1007-9327/full/v15/i33/4183.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.4183
| n | S100A2 mRNA | S100A2 protein | |||
| Negative | Positive | Negative | Positive | ||
| Gender | |||||
| Male | 31 | 7 | 24 (77.4) | 8 | 23 (74.2) |
| Female | 9 | 2 | 7 (77.8) | 3 | 6 (66.7) |
| Age (yr) | |||||
| < 62 | 21 | 4 | 17 (81) | 4 | 17 (81) |
| ≥ 62 | 19 | 5 | 14 (73.7) | 7 | 12 (63.2) |
| Clinical stage | |||||
| I and II | 31 | 5 | 26 (83.9) | 6 | 25 (80.7) |
| III and IV | 9 | 4 | 5 (55.6) | 5 | 4 (44.4) |
| Tumor location | |||||
| Upper and middle segment | 23 | 3 | 20 (87) | 5 | 18 (78.3) |
| Inferior segment | 17 | 6 | 11 (64.7) | 6 | 11 (64.7) |
| Tumor size (cm) | |||||
| < 3.5 | 16 | 3 | 13 (81.3) | 4 | 12 (75) |
| ≥ 3.5 | 24 | 6 | 18 (75) | 7 | 17 (70.8) |
| Depth of tumor invasion | |||||
| Not to serosa | 20 | 4 | 16 (80) | 4 | 16 (80) |
| To serosa | 20 | 5 | 15 (75) | 7 | 13 (75) |
| Degree of differentiation | |||||
| Well | 15 | 1 | 14 (93.3) | 2 | 13 (86.7) |
| Moderately | 14 | 2 | 12 (85.7) | 2 | 12 (85.7) |
| Poorly | 11 | 6 | 5 (45.5)a | 7 | 4 (36.4)b |
| Lymph node metastasis | |||||
| Negative | 25 | 2 | 23 (92) | 2 | 23 (92) |
| Positive | 15 | 7 | 8 (53.3)d | 9 | 6 (40)d |
| S100A2 protein | S100A2 mRNA | Total | |
| + | - | ||
| + | 27 | 2 | 29 |
| - | 4 | 7 | 11 |
| Total | 31 | 9 | 40 |
| p63 protein | S100A2 protein | Total | |
| + | - | ||
| + | 11 | 10 | 21 |
| - | 18 | 1 | 19 |
| Total | 29 | 11 | 40 |
Esophageal carcinoma (EC) is one of the most common malignant diseases in China and has a poor survival rate. The carcinogenesis and development of EC is a complex process referring to multiple factors, stages and genovariations, and to numerous changes in genes and proteins at the molecular level. However, the biological roles of these changes in esophageal carcinogenesis are still vague. Up to now, no specific tumor marker for EC has been identified, and there are no biomarkers for use in screening, early diagnosis and judgment of the biological behaviour of EC.
As a tumor suppressor gene, S100A2 is located on human chromosome 1q21[1] and its encoding protein is a calcium-binding protein constituted by 97 amino acids[2]. As a member of the S100 family, S100A2 is individual compared with other members of this family. Firstly, S100A2 protein is predominantly located in the nucleus rather than the cytosol like other S100 proteins. Moreover, S100A2 is down-regulated in several tumors and may play a role in inhibiting tumor initiation or in suppressing tumor cell growth, whereas other members of the S100 family are up-regulated in tumors and may play a promotional role in carcinogenesis. Therefore, S100A2 is considered a candidate tumor-suppressor gene[3].
p63 gene, a new member of the p53 gene family, was identified and named in 1998 by Yang et al[4]. p63 gene is located on human chromosome 3q27-29 and expresses at least six protein isoforms, which can be divided into two groups -TAp63 and ΔNp63[5]. Among them, the transcription level of ΔNp63α is the highest. TAp63 is able to activate the transcription of specific target genes and induce cell cycle arrest and apoptosis. Similar to p53, TAp63 has anti-oncogene activity. ΔNp63 is unable to activate transcription and inhibit transcription activation by both p53 and TAp63. ΔNp63 has anti-apoptosis and proto-oncogene activity[6,7].
Normally, p63 is mainly expressed in the basal lamina of many epithelial tissues and plays an important role in initiating epithelial stratification during development and in maintaining the proliferative potential of basal keratinocytes in mature epidermis. Recently, p63 gene has been studied in the fields of tumorigenesis, cell apoptosis and tissue growth.
At present, few studies have been performed on the expression and relationship of S100A2 and p63 in EC. In this study, in situ hybridization (ISH) used to identify S100A2 mRNA and immunohistochemical staining (S-P method) used to identify S100A2 and p63 protein, were performed in 40 samples of esophageal squamous cell carcinoma (ESCC) and 40 samples of normal esophageal mucosa. The purpose of this study was to investigate the expression of S100A2 mRNA, S100A2 protein and p63 protein in EC, and their relationship with clinical pathological features, and to explore their roles in the carcinogenesis and prognosis of EC.
Forty specimens of ESCC and matched adjacent normal mucosa were obtained from the Department of Pathology, First Affiliated Hospital, Anhui Medical University between 2005 and 2006. None of the patients had been treated with radiotherapy or chemotherapy prior to surgery. Samples were taken from tumor tissue without hemorrhage or putrescence, whereas the matched normal mucosa samples were taken from the surgical cutting edge, which was approximately 3-5 cm from the cancerous lesion. The clinical diagnosis in all 40 patients was confirmed by histological examination after surgery.
ISH reagents were purchased from Boster Co. (Wuhan, China) and Zhong Shan Co. (Beijing, China). A digoxigenin-labeled oligonucleotide probe was used. The probe sequence of S100A2 is described as: 5'-TGATGTGCAGTTCTCTGGAGCAGGCGCTGGCTGTG-3'; 5'-ACTGTCATGTGCAATGACTTCTTCCAGGGCTGCCC-3'. ISH for S100A2 was performed as follows: The tissue sections were treated with 3% hydrogen peroxide and 10% pepsin (diluted with 3% citric acid), respectively, after deparaffinization and rehydration. The sections were pre-hybridized at 37°C for 4 h with a prehybridization solution (Boster Co., China). Next, the sections were incubated in 100 μL hybridization solution/section containing 1 μL of denatured probe and 400 μL dilution of oligonucleotide probe (Boster Co., China) at 43°C for 16-20 h. The slides were washed at 37°C in 2 × SSC (5 min, three times), 0.5 × SSC (5 min, three times) and 0.2 × SSC (5 min, three times), respectively. During the color reaction procedure, the slides were incubated in sheep serum at 37°C for 30 min and then incubated with mouse antidigoxigenin antibody at 37°C for 60 min. After washing with PBS, the color was developed in DAB (3,3’-diaminobenzidine) (Zhongshan Co., China) for 15-30 min identified by occasional observation. Counterstaining of slides was then conducted with hematoxylin followed by a sealing procedure with neural gum.
S100A2 polyclonal antibody was purchased from Neomarker Co. (USA) and was used at a concentration of 1:100, and p63 monoclonal antibody was purchased from Maixin Co. (China). Immunohistochemistry for S100A2 and p63 were performed as follows: Deparaffinization and rehydration of sections (3 × 3 min with xylene; 3 × 2 min with 100% ethanol; 2 min with 95% ethanol, 2 min with 75% ethanol, and 2 × 1 min with distilled water), was performed using microwave repaired antigen. The samples were then washed 3 × 5 min with PBS. Endogenous peroxidases were blocked by soaking the slides in a solution of 3% H2O2 for 15 min at room temperature (RT). The samples were then washed 3 × 5 min with PBS, and 50 μL of nonimmune animal blood serum was added to each section for 15 min at RT. The samples were shaken and excess PBS was wiped off. Primary antibody (50 μL) was added to each section immediately, and incubated overnight at 4°C in a humidified chamber. The slides were washed 3 × 10 min in PBS. Fifty microliters of biotin labeling secondary antibody was added to each section for 15 min at 37°C, then the slides were washed 3 × 5 min with PBS. (ABC was made according to the Vector protocol 30 min before use (mix 5 mL PBS with two drops of solution A and two drops of solution B). The samples were incubated for 15 min at RT, then washed 3 × 5 min in PBS. Freshly prepared DAB (100 μL) was added to the slides and the color change was observed approximately 5-20 min later. Coloration was stopped by flushing with water. The slides were then counterstained with hematoxylin followed by a sealing procedure with neural gum.
Normal epithelium showing strong S100A2 or a p63-positive section served as the positive control, and that without the antibody section served as the negative control.
S100A2 mRNA and S100A2 protein were expressed in the nucleus or the nucleus and cytoplasm. The percentage of positive cells in each high power field was noted, and less than 10% of positive cells was considered negative and greater than 10% was considered positive. p63 protein was expressed in the nucleus. The grading of positive cells was as follows: no p63-positive cells was characterized as negative, less than 25% was weakly positive (+), 25%-75% was moderately positive (++), and greater than 75% was strongly positive (+++). The percentage of staining was estimated by two independent pathologists, respectively.
All data were analyzed with SPSS 13.0 software. Pearson’s χ2 test was used for data measurement. Using the Monte Carlo simulation method, the exact probability value was calculated for the data where the theoretical frequency was less than five. Non-parametric Spearman rank correlation analysis was used for correlation analysis of ranked data.
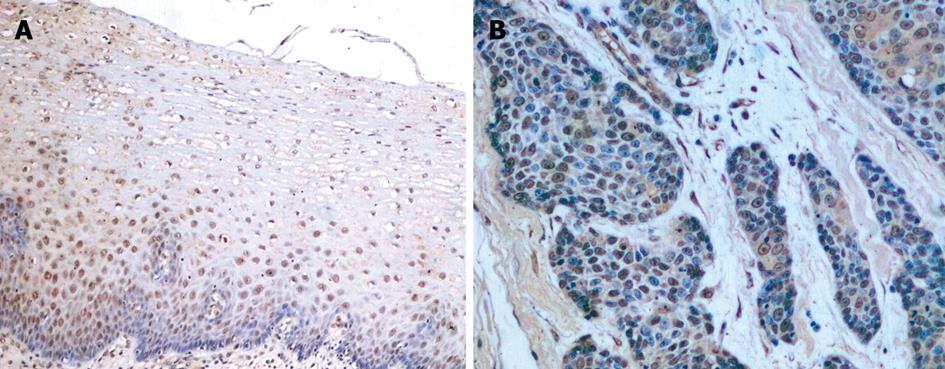
S100A2 mRNA and S100A2 protein were expressed in the nucleus or the nucleus and cytoplasm (Figure 1A and B, Figure 2A and B). All 40 samples of normal esophageal mucosa positively expressed S100A2 mRNA and S100A2 protein. In 40 ESCC samples, 31 positively expressed S100A2 mRNA, and 29 expressed S100A2 protein. The positive rates of S100A2 mRNA and S100A2 protein in ESCC (77.5% and 72.5%, respectively) were significantly lower than that in normal mucosa (100% and 100%, respectively) (P < 0.01) (Table 1).
S100A2 mRNA was positive in 93.3% (14/15), 85.7% (12/14), and 45.5% (5/11) in the well differentiated, moderately differentiated, and poorly differentiated groups, respectively. The differences were significant among the three groups (P < 0.05). Moreover, the expression level of S100A2 mRNA was significantly higher in the well and moderately differentiated groups than that in the poorly differentiated group (P < 0.05). The expression of S100A2 was positively correlated with node metastasis (P < 0.01). S100A2 protein was positive in 86.7% (13/15), 85.7% (12/14), and 36.4% (4/11) in the well differentiated, moderately differentiated, and poorly differentiated groups, respectively. The expression level of S100A2 protein was related to differentiation and lymph node metastasis. The expression of S100A2 mRNA and S100A2 protein was not correlated with gender, age, clinical stage, tumor location, tumor size, and depth of tumor invasion (P > 0.05) (Table 2).
A positive correlation was found between the expression of S100A2 mRNA and S100A2 protein (r = 0.607, P < 0.001). Of 40 ESCC samples, 27 expressed S100A2 mRNA and S100A2 protein at the same time, and seven cases did not express these parameters at the same time. Four cases expressed S100A2 mRNA but not S100A2 protein. Two cases did not express S100A2 mRNA but expressed S100A2 protein (Table 3).
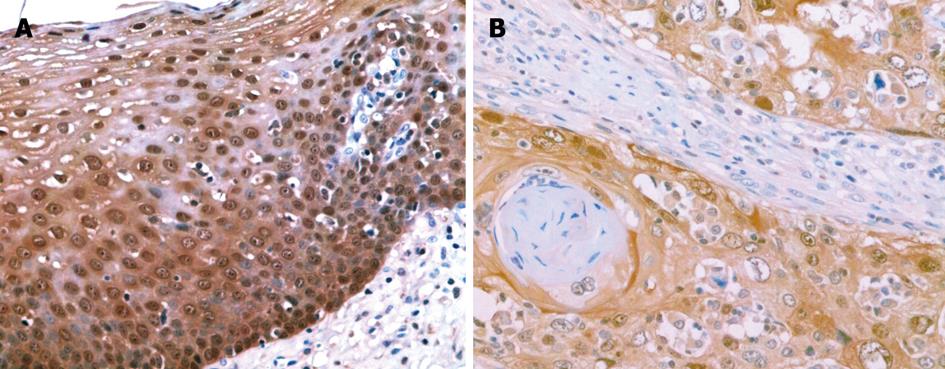
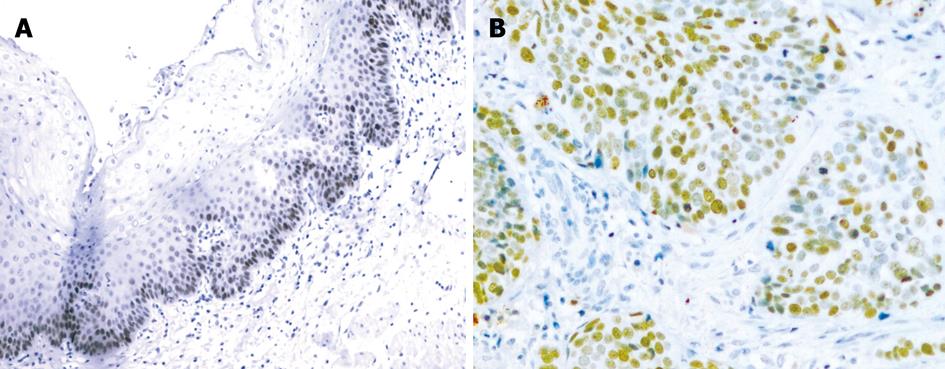
p63 protein was expressed in the nucleus (Figure 3A and B). In 40 samples of normal esophageal mucosa, 10 expressed p63 but none were strongly positive. In 40 ESCC samples, 21 expressed p63. Among these, 15 cases were strongly positive. p63 protein was positive in 52.5% and strongly positive in 37.5% of ESCC samples, and was higher than that in normal mucosa (25.0%) (P < 0.05, P < 0.001) (Table 4).
The expression of p63 protein was not correlated with gender, age, clinical stage, tumor location, tumor size, depth of tumor invasion, degree of differentiation or lymph node metastasis (P > 0.05).
A negative correlation was found between the expression of S100A2 protein and p63 protein (r = -0.474, P < 0.01). Of 40 ESCC samples, 11 expressed S100A2 and p63 at the same time, and 1 did not express these parameters at the same time. 18 ESCC samples were S100A2-positive and p63-negative. 10 cases were S100A2-negative and p63-positive (Table 5).
Up to now, 21 genes encoding S100 calcium-binding proteins of the EF-hand type have been identified. Calcium ion (Ca2+) plays an important role in the regulation of a number of cellular processes. The second messenger role of Ca2+ is mediated, at least in part, by calcium-binding proteins which contain the EF-hand motif. Through modulating Ca2+ and interacting with the target proteins, S100 proteins have a multitude of biological functions in vivo. Many S100 genes were reported to be clustered in the epidermal differentiation complex in chromosome 1q21[8-12]. This region is involved in epidermal differentiation and proliferation and is also frequently rearranged in tumors[13]. Studies have shown that the S100 gene deregulated expression in human diseases, especially in tumors. Nowadays, S100 protein antibodies have widespread application for tumor diagnosis by immunohistochemistry.
As a member of the S100 gene family, S100A2 is significantly downregulated in several malignant tumors, such as breast cancer[14], melanoma[15], prostatic carcinoma[16], and pulmonary carcinoma[17]. Moreover, S100A2 may be closely associated with the development and prognosis of tumors[18].
Using semiquantitative reverse transcription-polymerase chain reaction (RT-PCR), Ji et al[19] examined the differential expression of S100A2 and another 15 S100 genes in 62 cases of ESCC vs the corresponding normal esophageal mucosa. Their results showed that the S100A2 gene was significantly downregulated (P < 0.05) in ESCC vs normal esophageal mucosa. Moreover, the deregulation of S100A2 gene was significantly correlated with the deregulation of S100A8, S100A14 and S100P. Kyriazanos et al[20] examined the clinical significance of S100A2 expression in 116 resected specimens of ESCC using immunohistochemistry. Their results showed that S100A2 was positive in 49 cases (42.2%) and its expression was significantly higher in large and well differentiated tumors. Lymph node-positive tumors had a lower expression of S100A2 protein in comparison to the corresponding lymph node-negative equivalents in each of the T stages, but the difference was statistically significant only for T1b tumors. S100A2 status became an independent predictor of patient survival in lymph node-negative cases. Node-negative ESCC patients without S100A2 expression may be a high-risk group with poor survival and will need further attention to design appropriate adjuvant therapy.
We used ISH for the first time to detect the expression of S100A2 mRNA and used immunohistochemical staining for S100A2 protein in 40 ESCC samples and 40 samples of normal esophageal mucosa. Our results showed that S100A2 mRNA and S100A2 protein although positive in ESCC were both lower than that in normal mucosa (P < 0.01). This indicated that S100A2 was downregulated in ESCC and that the S100A2 gene is concerned with the carcinogenesis of EC. A positive correlation was found between the expression of S100A2 mRNA and S100A2 protein (P < 0.001), which indicated that S100A2 protein expression is regulated mainly at the transcriptional level, and the decrease in S100A2 protein expression corresponded to the decrease in transcriptional activity. In addition, our findings on S100A2 mRNA are consistent with those of Ji et al[19]. We suggest that the positive rate in our study is higher than that in the study by Ji et al[19] due to the difference in case number (40 vs 62) and method (ISH vs RT-PCR).
Furthermore, we analyzed the relationship between S100A2 positive expression and the clinical pathological features of ESCC. We found that the expression level of S100A2 mRNA and S100A2 protein were both significantly higher in the well and moderately differentiated groups than in the poorly differentiated group (P < 0.05). These differences were also significant between the lymph node-positive group and the lymph node-negative group (P < 0.01), which is roughly consistent with the findings of Kyriazanos et al[20]. These results indicate that S100A2 plays an important role in tumor cell differentiation, and that S100A2 might be an important biomarker in the biological behaviour of EC.
A study on the course of EC, found that p63 gene and its encoding protein played important roles in the early period of the physiological and pathological course of esophageal mucosa, compared with mutation of p53 which occurs in the last stage of cancerization lineage from metaplasia, atypical hyperplasia to adenocarcinoma. Glickman et al[21] detected p63 protein in EC by immunohistochemical staining. Their results showed that p63 protein was highly expressed in ESCC, but was not expressed in adenocarcinoma of the esophagus and colorectal cancer. This indicated that p63 gene is upregulated in ESCC and is concerned with the development of ESCC.
We used immunohistochemical staining for p63 protein in 40 ESCC samples and 40 samples of normal esophageal mucosa. Our results showed that p63 protein in normal esophageal mucosa was positive in 25.0%, and the expression was weakly positive and localization in the basal cells or the bottom of the prickle cell layer of normal esophageal mucosa. This supports the view that p63 is mainly expressed in the corpus of many epithelial tissues, and plays an important role in the abstraction, differentiation and morphogenesis of many epithelial tissues[22]. Moreover, our results showed that p63 protein has a high expression rate in the tissue of EC, which indicates that p63 is closely related to the carcinogenesis of EC. In contrast, we found that the expression of p63 protein was not correlated with the clinical pathological features of ESCC.
Our study showed that the expression of S100A2 protein was reduced and the expression of p63 protein was increased, and a negative correlation was observed between them (P < 0.01). This indicated that S100A2 protein and p63 protein may both play important roles in the carcinogenesis of EC. An investigation into the combined expression of S100A2 and p63 may be useful in early diagnosis and in evaluating the prognosis of ESCC.
Esophageal carcinoma (EC) is one of the most common malignant diseases in China and has a poor survival rate. Up to now, no specific tumor marker of EC has been identified. Recently, S100A2 has been considered a candidate tumor-suppressor gene, and p63 gene has been studied in the fields of tumorigenesis, cell apoptosis and tissue growth.
S100A2 is down-regulated in several tumors and may play a role in inhibiting tumor initiation or in suppressing tumor cell growth. Therefore, S100A2 has been considered a candidate tumor-suppressor gene. p63 gene, a new member of the p53 gene family, is normally expressed in the basal lamina of many epithelial tissues and plays an important role in initiating epithelial stratification during development and in maintaining proliferative potential of basal keratinocytes in mature epidermis. Recently, p63 gene has been studied in the fields of tumorigenesis, cell apoptosis and tissue growth.
At present, few studies have been carried out on the expression and the relationship of S100A2 and p63 in EC. In this study, in situ hybridization (ISH) to identify S100A2 mRNA and immunohistochemical staining (S-P method) to identify S100A2 and p63 protein were performed in ESCC and normal esophageal mucosa samples.
S100A2 gene is located on human chromosome 1q21 and its encoding protein is a calcium-binding protein constituted by 97 amino acids. S100A2 protein is predominantly located in the nucleus rather than the cytosol like other S100 proteins. S100A2 is down-regulated in several tumors. Therefore, S100A2 is considered a candidate tumor-suppressor gene.
The study investigated S100A2 and p63 expression in EC tissue samples and paired normal mucosa samples. S100A2 expression was analysed both on the mRNA and protein level by ISH and IH, respectively, p63 expression was determined by IH. The authors described a decreased expression of S100A2 and an increased expression of p63 in tumor samples. Statistical analysis revealed an inverse correlation. This is a clear strength of this study.
| 1. | Engelkamp D, Schafer BW, Mattei MG, Erne P, Heizmann CW. Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci USA. 1993;90:6547-6551. |
| 2. | Nakayama S, Kretsinger RH. Evolution of the EF-hand family of proteins. Annu Rev Biophys Biomol Struct. 1994;23:473-507. |
| 3. | Wicki R, Franz C, Scholl FA, Heizmann CW, Schafer BW. Repression of the candidate tumor suppressor gene S100A2 in breast cancer is mediated by site-specific hypermethylation. Cell Calcium. 1997;22:243-254. |
| 4. | Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305-316. |
| 5. | Tannapfel A, Schmelzer S, Benicke M, Klimpfinger M, Kohlhaw K, Mossner J, Engeland K, Wittekind C. Expression of the p53 homologues p63 and p73 in multiple simultaneous gastric cancer. J Pathol. 2001;195:163-170. |
| 6. | Sasaki Y, Morimoto I, Ishida S, Yamashita T, Imai K, Tokino T. Adenovirus-mediated transfer of the p53 family genes, p73 and p51/p63 induces cell cycle arrest and apoptosis in colorectal cancer cell lines: potential application to gene therapy of colorectal cancer. Gene Ther. 2001;8:1401-1408. |
| 7. | Ito Y, Takeda T, Wakasa K, Tsujimoto M, Sakon M, Matsuura N. Expression of p73 and p63 proteins in pancreatic adenocarcinoma: p73 overexpression is inversely correlated with biological aggressiveness. Int J Mol Med. 2001;8:67-71. |
| 8. | Gendler SJ, Cohen EP, Craston A, Duhig T, Johnstone G, Barnes D. The locus of the polymorphic epithelial mucin (PEM) tumour antigen on chromosome 1q21 shows a high frequency of alteration in primary human breast tumours. Int J Cancer. 1990;45:431-435. |
| 9. | Schafer BW, Wicki R, Engelkamp D, Mattei MG, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics. 1995;25:638-643. |
| 10. | Weterman MA, Wilbrink M, Dijkhuizen T, van den Berg E, Geurts van Kessel A. Fine mapping of the 1q21 breakpoint of the papillary renal cell carcinoma-associated (X;1) translocation. Hum Genet. 1996;98:16-21. |
| 11. | Pietas A, Schluns K, Marenholz I, Schafer BW, Heizmann CW, Petersen I. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics. 2002;79:513-522. |
| 12. | Watson PH, Leygue ER, Murphy LC. Psoriasin (S100A7). Int J Biochem Cell Biol. 1998;30:567-571. |
| 13. | Lioumi M, Olavesen MG, Nizetic D, Ragoussis J. High-resolution YAC fragmentation map of 1q21. Genomics. 1998;49:200-208. |
| 14. | Liu D, Rudland PS, Sibson DR, Platt-Higgins A, Barraclough R. Expression of calcium-binding protein S100A2 in breast lesions. Br J Cancer. 2000;83:1473-1479. |
| 15. | Maelandsmo GM, Florenes VA, Mellingsaeter T, Hovig E, Kerbel RS, Fodstad O. Differential expression patterns of S100A2, S100A4 and S100A6 during progression of human malignant melanoma. Int J Cancer. 1997;74:464-469. |
| 16. | Gupta S, Hussain T, MacLennan GT, Fu P, Patel J, Mukhtar H. Differential expression of S100A2 and S100A4 during progression of human prostate adenocarcinoma. J Clin Oncol. 2003;21:106-112. |
| 17. | Matsubara D, Niki T, Ishikawa S, Goto A, Ohara E, Yokomizo T, Heizmann CW, Aburatani H, Moriyama S, Moriyama H. Differential expression of S100A2 and S100A4 in lung adenocarcinomas: clinicopathological significance, relationship to p53 and identification of their target genes. Cancer Sci. 2005;96:844-857. |
| 18. | Lauriola L, Michetti F, Maggiano N, Galli J, Cadoni G, Schafer BW, Heizmann CW, Ranelletti FO. Prognostic significance of the Ca(2+) binding protein S100A2 in laryngeal squamous-cell carcinoma. Int J Cancer. 2000;89:345-349. |
| 19. | Ji J, Zhao L, Wang X, Zhou C, Ding F, Su L, Zhang C, Mao X, Wu M, Liu Z. Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:480-486. |
| 20. | Kyriazanos ID, Tachibana M, Dhar DK, Shibakita M, Ono T, Kohno H, Nagasue N. Expression and prognostic significance of S100A2 protein in squamous cell carcinoma of the esophagus. Oncol Rep. 2002;9:503-510. |
| 21. | Glickman JN, Yang A, Shahsafaei A, McKeon F, Odze RD. Expression of p53-related protein p63 in the gastrointestinal tract and in esophageal metaplastic and neoplastic disorders. Hum Pathol. 2001;32:1157-1165. |