INTRODUCTION
Many eukaryotes require iron for critical biological processes[12]. However, excess iron is toxic, causing lipid peroxidation and oxidative stress[3]. Due to it being a “double edged sword”, iron uptake and transport must therefore be tightly regulated[4–7]. Hepcidin, a circulatory peptide synthesized by the liver, performs this important task by regulating iron transport in different parts of the body including duodenum, spleen and bone marrow[89]. Alcohol has been shown to suppress hepcidin expression in the liver leading to an increase in intestinal iron transport[10–15]. Both iron and alcohol act synergistically to cause liver injury. Disruption of hepcidin synthesis by alcohol may therefore be one of the “second hit” mechanisms leading to the progression of alcoholic liver disease.
ALCOHOL-INDUCED DISTURBANCES IN IRON METABOLISM
Alcohol consumption has long been associated with changes in iron homeostasis. The reported changes range from anemia to iron overload[16–22]. Blood loss is common in patients with alcoholic cirrhosis[23]. Anemia and iron deficiency can be due to gastrointestinal blood loss arising from the complications of alcohol abuse. Nutritional deficiencies, such as folate deficiency, can also be a common cause of anemia[24–27]. Megaloblastic and sideroblastic anemias, macrocytosis of alcoholism, elevated mean corpuscular volume, and iron deficiency have been reported to be common among hospitalized chronic alcoholics[2327–30]. However, it has been demonstrated that anemia and megaloblastic and sideroblastic changes do not occur if an adequate diet is provided during chronic alcohol administration to human volunteers[2531]. This has also been shown to be the case for relatively well-nourished alcoholics[25263233]. These findings therefore suggest that alcohol by itself does not induce iron deficiency or anemia.
In fact, wine has a high iron content and increases iron absorption[34]. Decreasing the alcohol content of red wine has been reported to significantly reduce the absorption of non-heme iron in human subjects[35]. Beer consumption has been suggested to have a more significant effect on serum ferritin levels than wine and spirits, both in males and females[18]. Alcohol consumption (up to 2 alcoholic drinks per day) has also been shown to exert a protective effect by reducing the risk for iron deficiency anemia in adult participants of the Third National Health and Nutrition Examination Survey[36]. On the other hand, heavy alcohol consumption (more than two alcoholic drinks per day) elevates the risk of iron overload[36]. In both male and female adolescents (aged 16-19 years), who participated in the First National Health and Nutrition Examination Survey, serum iron concentration was significantly related to drinking frequency[17]. Alcohol consumption was associated with elevated serum iron concentration in both male and female adolescents. Interestingly, male adolescents also exhibited increased transferrin saturation and hemoglobin concentration[17]. Drinking frequency was unrelated to dietary iron intake, poverty index or race[17]. A Danish population survey also reported a correlation between alcohol intake and elevated serum ferritin in healthy adult males and females (aged 30-60 years)[37]. African Americans who consume alcohol (4 drinks per day) exhibit a higher prevalence of ferroportin Q248H allele, which is implicated in iron accumulation, and alcohol has been suggested to contribute to higher body iron stores in this population[38]. Alcohol elevates iron absorption and patients with alcoholic cirrhosis often exhibit elevated liver iron content[3940]. In vivo whole-body retention studies have demonstrated a two-fold increase in intestinal iron absorption in chronic alcoholics[41]. Alcoholic cirrhosis patients with higher liver iron content display increased mortality rates[42]. Experimental animal models of prolonged alcohol exposure also display increased liver iron deposition and hepatocellular injury[1943]. In mild cases of alcoholic liver disease (ALD), iron has been reported to accumulate in hepatocytes[44]. However, in patients with advanced ALD, both parenchymal cells and Kupffer cells exhibit iron staining[20].
In some alcoholics, the elevated serum iron indices have been reported to significantly decrease within 2-6 wk of abstinence, suggesting a direct role for alcohol consumption[45]. However, the mechanisms by which alcohol disrupts iron metabolism are unclear. Recent studies indicate a role for hepcidin in this process.
HEPCIDIN
The discovery of the circulating peptide hormone hepcidin has revolutionized our understanding of iron metabolism. Hepcidin sequence is highly conserved between species, from fish to mammals. Hepcidin was first isolated from human urine and serum as short cysteine-rich peptide forms[4647]. Hepcidin is homologous to the members of the defensin family of antimicrobial peptides and has been reported to exhibit antimicrobial activity in vitro[46]. It is the studies with transgenic mice that have highlighted the importance of this protein in the regulation of iron metabolism. Namely, hepcidin knockout mice, deficient in the expression of hepcidin, develop severe iron overload, whereas mice overexpressing hepcidin display iron deficiency and anemia[4849]. Moreover, the synthesis of hepcidin has also been shown to be responsive to the body iron levels. Iron overload state induces hepcidin synthesis whereas iron deficiency and anemia have the opposite effect[5051].
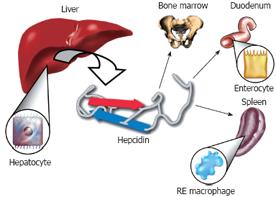
Liver is the main site of hepcidin synthesis in the body[50]. It is synthesized as an 84 amino acid precursor protein and is subsequently cleaved into the biologically active 25 amino acid peptide form[895052]. In the liver, hepcidin is primarily expressed in the hepatocytes (Figure 1). Although to a much lesser extent, other cells of the liver and other organs also display hepcidin expression. The exciting discovery of the interaction between hepcidin and the iron exporter protein ferroportin was pivotal to our understanding of how hepcidin regulates iron metabolism[5354]. The binding of hepcidin to ferroportin induces the internalization and degradation of the ferroportin protein, which in turn inhibits iron from being transported into the circulation. This is how hepcidin accomplishes the regulation of iron mobilization between distant locations (i.e. duodenum, bone marrow, reticuloendothelial macrophages) in the body (Figure 1)[8].
Figure 1 The Iron regulatory hormone hepcidin.
Hepcidin is synthesized in the hepatocytes of the liver as an 84 amino acid precursor protein. It is subsequently cleaved into the 25 amino acid biologically active peptide form and is released into the circulation. Hepcidin plays a central role in the regulation of iron metabolism by inhibiting the release of iron from the enterocytes of the duodenum and from reticuloendothelial macrophages. Hepcidin blocks the export of iron from these cells by binding to the iron exporter protein, ferroportin, which induces the internalization and degradation of ferroportin protein. As a soluble mediator, hepcidin establishes the cross-talk between distant organs in the body in order to maintain iron homeostasis.
The synthesis of hepcidin in the liver is modulated by numerous upstream regulators[12]. Transferrin receptor 2, the genetic hemochromatosis gene product, Hfe, and the juvenile hemochromatosis gene product, Hjv, are positive regulators of hepcidin expression[55–62]. Mutations in these genes result in an increase in body iron stores (i.e. hemochromatosis) due to the down-regulation of hepcidin expression. Recently, a novel candidate TMPRSS6, a transmembrane serine protease also known as matriptase 2, has been identified. Interestingly, TMPRSS6 is the first identified negative regulator of liver hepcidin expression. In other words, TMPRSS6 keeps hepcidin expression under control in the liver[6364]. Accordingly, patients expressing TMPRSS6 mutations exhibit iron-refractory iron deficiency anemia due to elevated hepcidin production[63]. Moreover, the splicing defects in the TMPRSS6 gene lead to the mask phenotype, a recessive, chemically induced mutant mouse phenotype[64]. This phenotype results from the reduced absorption of dietary iron due to high levels of hepcidin expression[64]. However, the signaling mechanisms involved in these regulatory processes (negative or positive) are as yet unknown.
KUPFFER CELLS, ALCOHOL AND HEPCIDIN
Liver macrophages (Kupffer cells) also play a pivotal role in the progression of ALD. Alcoholics have elevated levels of lipopolysaccharide (LPS) in their circulation, which is believed to prime the macrophages leading to the release of proinflammatory cytokines, particularly TNF-α, and reactive oxygen species[65–67]. The importance of Kupffer cells in ALD has been demonstrated in studies where their depletion or inactivation blocked alcohol-induced symptoms, including inflammation, fatty liver and necrosis, in experimental animal models of chronic alcohol exposure[6869].
In advanced forms of ALD and in experimental animal models of ALD, besides parenchymal cells, Kupffer cells also exhibit iron accumulation[207071]. Reticuloendothelial (RE) macrophages play a major role in iron trafficking and recycling to meet changes in the body’s iron demand[72]. Like the parenchymal cells of the liver, the RE system also stores large amounts of iron bound to the storage protein, ferritin. RE macrophages acquire iron mainly by phagocytosing senescent red blood cells. However, macrophages also express the divalent metal transporter1 (DMT1), natural resistance associated macrophage protein1 (Nramp1), hemoglobin scavenger receptor (CD163) and transferrin receptor1 (TrfR1), all of which are involved in iron uptake and transport[72–75]. The livers from experimental animal models of ALD and Japanese patients with ALD have been reported to exhibit significantly more TrfR1 mRNA or protein expression, respectively, compared to control animals or healthy human subjects[7677]. Macrophages can take up transferrin-bound iron in vitro. However, a significant uptake of transferrin-bound iron by macrophages has not been observed in vivo in humans[7278]. Although an increase in TrfR1 mRNA expression was observed in Kupffer cells of experimental animals, the increase in TrfR1 protein expression in Japanese patients with ALD was observed mainly in the hepatocytes[207677]. It should also be noted that mice with disrupted TrfR1 gene, deficient in the global expression of TrfR1 receptor, display abnormalities only in erythropoiesis and neurologic development[79]. The Kupffer cells of rats exposed to chronic alcohol have also been shown to display an increase in mRNA levels of Hfe, the gene for genetic hemochromatosis[77]. Hfe gene product does not bind iron and is not directly involved in iron uptake. However, it is involved in the regulation of iron metabolism as shown by the fact that mice deficient in Hfe expression develop iron overload[8081]. Hfe is believed to achieve this by modulating the expression and iron responsiveness of hepcidin in the liver[575882]. On the other hand, the iron release from macrophages is regulated by the iron exporter protein ferroportin[8384]. The multicopper ferroxidase, ceruloplasmin may also play a role in iron efflux in macrophages[85–87]. The Kupffer cells of animals subjected to alcohol exposure have been shown to display an increase in both ferroportin mRNA and protein, indicative of increased iron export[77]. Of note, hepcidin binds to ferroportin and induces its internalization and degradation[5354]. The increase in ferroportin expression in the Kupffer cells of these animals may be due to suppressed hepcidin expression. Alcohol has been shown to down-regulate hepcidin expression both in animal models of ALD and in patients with ALD[11–15]. However, Xiong et al[77] have reported no change in plasma prohepcidin levels and a decrease in hepcidin mRNA levels in Kupffer cells in their experimental model.
Iron has been reported to prime Kupffer cells for alcoholic liver injury in rats exposed to chronic intragastric alcohol infusion[70], The proinflammatory cytokine TNF-α is a key player in alcohol-induced liver injury[88–90]. Iron has been shown to activate the transcription factor, NF-κB in hepatic macrophages and induce the synthesis and release of TNF-α[7091]. Following erythrophagocytosis, Kupffer cells have been shown to display elevated levels of LPS-induced NF-κB activation[70]. Treatment of cultured Kupffer cells in vitro and ex vivo with iron chelators blocked both NF-κB activation and TNF-α synthesis[7071]. Proinflammatory cytokines (IL-1, IL-6, TNF-α) have been suggested to increase iron uptake into the monocytes of patients with rheumatoid arthritis[92]. It is therefore possible that alcohol and/or iron-induced release of TNF-α may serve as a feedback loop for further iron uptake into the Kupffer cells. On the other hand, Olynyk et al[93] have reported that deposition of iron in Kupffer cells impairs LPS-induced proinflammatory cytokine production in these cells. Xiong et al[77] have reported that the intracellular labile non-heme iron pool is involved in peroxynitrite or LPS-mediated activation of NF-κB and TNF-α release in Kupffer cells isolated from experimental animal models of chronic alcohol exposure. The intracellular labile iron pool is a transitory, free (non-ferritin-bound) pool of iron, which is chelatable by commonly used iron chelators. This pool of iron contributes to the generation of reactive oxygen species but its biological relevance is unclear[94]. Transient changes in the dynamics of the labile iron pool is not only affected by increased iron uptake but also by prooxidant chemicals, which induce the reductive release of iron from intracellular stores[9596]. Xiong et al[77] have demonstrated that iron dextran treatment of cultured Kupffer cells in vitro does not affect the basal TNF-α release. Interestingly, Kupffer cells isolated from mice two weeks after iron dextran injection, exhibited significant increases in basal TNF-α release (independent of peroxynitrite or LPS stimulation). These findings suggest that either other cells in the liver (besides Kupffer cells) or yet unknown signals contribute to this process in vivo. It is also possible that the discrepancy between in vitro and in vivo iron loading regarding basal TNF-α release from Kupffer cells may not be due to iron (iron loading) per se but rather due to the differences in the level of oxidative stress.
The synthesis of hepcidin in the liver is mediated by several stimuli including iron and inflammation. Kupffer cells have been shown not to play a role in the regulation of hepcidin expression by iron in vivo[9798]. The data in the literature regarding the involvement of Kupffer cells in the regulation of liver hepcidin expression by inflammation is contradictory[97–99]. Montosi et al[97] have reported a role for Kupffer cells in the regulation of hepcidin expression by inflammation in mice. On the other hand, Lou et al[98] and Theurl et al[99] have demonstrated that the depletion of Kupffer cells in mice does not abrogate the up-regulation of liver hepcidin expression by LPS.
The role of Kupffer cell activation and TNF-α signaling in the regulation of hepcidin expression by alcohol in vivo has also been reported[100]. The inactivation of Kupffer cells by gadolinium chloride in rats pair-fed with alcohol Lieber DeCarli diets for 6 wk or mice fed with ethanol in the drinking water for 1 wk did not reverse the alcohol-induced suppression of hepcidin expression in the liver[100]. Moreover, similar results were obtained when Kupffer cells were depleted by liposomes containing clodronate[100]. When phagocytosed by the Kupffer cells, clodronate released from the liposomes induces apoptosis and thereby depletes the Kupffer cell[101]. When co-cultured, Kupffer cells have been suggested to exert a negative effect on hepcidin synthesis in hepatocytes[99]. However, the depletion or inactivation of the Kupffer cells has been reported to not induce any significant changes in basal hepcidin expression levels in the livers of both control and alcohol-treated animals in vivo[100]. Interestingly, one week of ethanol treatment (in the drinking water) was sufficient to induce NF-κB activation and TNF-α and IL-6 release in mice, compared to control mice fed with plain water[100]. The neutralization of TNF-α inhibited the activation of NF-κB[100]. However, neither the neutralization of TNF-α nor the absence of TNF-α receptorIand II expression altered the effect of alcohol on hepcidin expression (i.e. down-regulation)[100]. These findings therefore strongly suggest that the activation of Kupffer cells and TNF-α signaling are not involved in the regulation of hepcidin expression by alcohol in vivo. These findings are also in agreement with Lou et al[98], who demonstrated that Kupffer cells are not required for the regulation of liver hepcidin expression by LPS-induced inflammation.
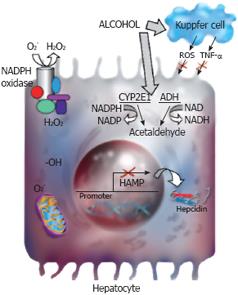
The suppression of hepcidin synthesis in the liver by alcohol occurs very early (within days) following alcohol exposure and involves hepatocytes, but not Kupffer cells (Figure 2)[12100]. It is noteworthy that hepatocytes are the main site of hepcidin synthesis in the liver[50]. Alcohol-induced oxidative stress in hepatocytes is one of the main mechanisms by which hepcidin expression in the liver is down-regulated by alcohol[12]. The decrease in liver hepcidin synthesis leads to an increase in intestinal iron transport and liver iron content[10–12]. Hence, alcohol dysregulates iron homeostasis by suppressing hepcidin expression in the liver. Although Kupffer cells are not involved in the initial stages of this process induced by alcohol, the increase in iron stores (due to low levels of hepcidin) will further activate Kupffer cells, and thereby lead to the release of proinflamatory cytokines. It is therefore possible that unlike LPS, the priming of Kupffer cells by iron[70] occurs in later stages of ALD and exacerbates the inflammatory processes by facilitating further release of TNF-α, and thereby contributing to liver injury.
Figure 2 Hepcidin and alcohol.
Alcohol is metabolized by alcohol dehydrogenase (ADH) and cytochrome P4502E1 (CYP2E1) in the liver. Alcohol-induced oxidative stress leads to the suppression of hepcidin promoter activity and hepcidin transcription in the liver. The parenchymal, but not the non-parenchymal cells of the liver are involved in the regulation of hepcidin transcription by alcohol-induced oxidative stress. The activation of CYP2E1 or NADPH oxidase and changes in mitochondrial functions are involved in alcohol-induced oxidative stress in hepatocytes. The role of these pathways in the regulation of hepcidin transcription by alcohol requires further investigation.
ALCOHOL-INDUCED OXIDATIVE STRESS AND HEPCIDIN
Alcohol metabolism generates reactive oxygen species and lipid peroxidation products (malondialdehyde, 4-hydroxynonenal) during the oxidation of ethanol by alcohol dehydrogenase and cytochrome P4502E1 to form acetaldehyde. Serum thioredoxin levels are also an indicator of oxidative stress and they have been reported to be significantly higher in patients with ALD compared to healthy subjects[102]. Due to its capacity to take part in the Fenton reaction as a transition metal, iron itself induces the generation of reactive oxygen species (superoxide, hydroxyl iron), which subsequently damage cellular membranes via lipid peroxidation[3103]. Iron and alcohol act as a deadly cocktail to exacerbate liver injury. Orally effective iron chelators have been reported to attenuate alcohol-induced hepatic lipid peroxidation in rats[104].
Alcohol-induced oxidative stress is involved in the suppression of hepcidin promoter activity and hepcidin transcription in the liver in vivo by inhibiting the DNA-binding activity of the transcription factor, C/EBP α[12]. Both Kupffer cells and hepatocytes are involved in alcohol-induced oxidative stress. However, a role for Kupffer cells in the regulation of hepcidin expression by alcohol in the liver has been excluded (Figure 2)[100]. This strongly suggests the involvement of hepatocytes in the regulation of hepcidin expression, and thereby iron metabolism by alcohol. Multiple pathways are involved in alcohol-induced oxidative stress in hepatocytes; redox state changes, the activation of CYP2E1 or NADPH oxidase and changes in mitochondrial functions[105–109]. Alcohol metabolism, which is accompanied by oxidation and reduction reactions, causes an imbalance in the redox state of the cell by generating excess NADH (reduced nicotinamide adenine dinucleotide). CYP2E1 enzyme, present in liver microsomes, has a high redox potential and is also metabolically active in the absence of alcohol[107]. It produces reactive oxygen species such as superoxide and hydrogen peroxide, which is increased by alcohol exposure leading to lipid peroxidation, oxidative stress and tissue injury. NADPH oxidase catalyzes the reduction of molecular oxygen to generate superoxide[110111]. The liver expresses both phagocytic and nonphagocytic isoforms of NADPH oxidase[112113]. Alcohol-induced liver pathology was not observed in mice with deficient NADPH oxidase activity suggesting a role for this enzyme in the progression of alcoholic liver disease[109]. Mitochondria play a key role both in iron biogenesis and alcohol metabolism[3108114]. Alcohol induces lesions in the proteins of the mitochondrial electron transport chain and decreases the rate of hepatic ATP synthesis[114–116]. This induces the transfer of unpaired electrons to molecular oxygen and elevates the production of reactive oxygen species. The involvement of these changes in the regulation of hepcidin expression in hepatocytes by alcohol needs further investigation (Figure 2).
Several factors including iron, viral infection, endotoxin and reactive oxygen species have been implicated to act as second hit factors in the progression of alcoholic liver disease. Of note, hepcidin expression in the liver is regulated by iron, alcohol, hepatitis C viral proteins, inflammation, hypoxia and oxidative stress[10–1248100117–120]. Nishina et al[119] have recently shown that hepatitis C viral protein-mediated oxidative stress suppresses hepcidin transcription by altering the activity of the transcription factor, C/EBP α. These findings are in agreement with our previously published results demonstrating a role for alcohol-mediated oxidative stress in the inhibition of C/EBP α activity and hepcidin transcription[1112]. Hepcidin may therefore act as a potential second hit factor in the progression of alcoholic liver disease.
CONCLUSION
Patients with alcoholic liver disease frequently display elevated iron stores. Recent reports by several groups strongly suggest that this may be a regulated mechanism rather than iron simply leaking out of injured intestinal cells. Hepcidin, an iron regulatory hormone synthesized in hepatocytes, plays a key role in the regulation of iron metabolism. Alcohol-mediated oxidative stress suppresses hepcidin expression in the liver leading to increased intestinal iron uptake and liver iron storage. Reduced hepcidin expression may be one of the mechanisms leading to elevated iron stores following alcohol exposure. Hepcidin may therefore act as a second hit in the progression of ALD. Both hepatocytes and Kupffer cells play a role in ALD. However, Kupffer cells have been reported to not be involved in the regulation of liver hepcidin expression by both acute and chronic alcohol exposure. Hepatocytes are the main site of alcohol metabolism in the liver leading to the generation of reactive oxygen species and the depletion of antioxidants. Alcohol-induced oxidative stress in hepatocytes is therefore one of the mechanisms leading to the suppression of hepcidin synthesis and thereby to iron overload, which in turn acts as a secondary risk factor in ALD. Accordingly, iron has been shown to prime Kupffer cells leading to the release of TNF-α in experimental animal models of ALD. The distruption of hepcidin synthesis and thereby iron metabolism by alcohol may have clinical relevance. Hepcidin therefore holds promise as an early diagnostic marker and as a candidate for therapeutic strategies for ALD.
Supported by Grants from the Alcoholic Beverage Medical Research Foundation, Redox Biology Center, University of Nebraska-Lincoln, 2P20RR017675 and NIH grant, R01AA017738-01 to DHF
Peer reviewer: Shivendra Shukla, Professor, Department of Medical Pharmacology and Physiology, University of Missouri School of Medicine, 1 Hospital Drive, M530 Medical Sciences Bldg., Columbia MO 65212, United States
S- Editor Li LF L- Editor Negro F E- Editor Ma WH