Published online Oct 7, 2008. doi: 10.3748/wjg.14.5657
Revised: August 26, 2008
Accepted: September 2, 2008
Published online: October 7, 2008
AIM: To isolate and identify differentially expressed proteins between cancer and normal tissues of gastric cancer by two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
METHODS: Soluble fraction proteins of gastric cancer tissues and paired normal tissues were separated by 2-DE. The differentially expressed proteins were selected and identified by MALDI-TOF-MS and database search.
RESULTS: 2-DE profiles with high resolution and reproducibility were obtained. Twenty-three protein spots were excised from sliver staining gel and digested in gel by trypsin, in which fifteen protein spots were identified successfully. Among the identified proteins, there were ten over-expressed and five under-expressed proteins in stomach cancer tissues compared with normal tissues.
CONCLUSION: In this study, the well-resolved, reproducible 2-DE patterns of human gastric cancer tissue and paired normal tissue were established and optimized and certain differentially-expressed proteins were identified. The combined use of 2-DE and MS provides an effective approach to screen for potential tumor markers.
- Citation: Li W, Li JF, Qu Y, Chen XH, Qin JM, Gu QL, Yan M, Zhu ZG, Liu BY. Comparative proteomics analysis of human gastric cancer. World J Gastroenterol 2008; 14(37): 5657-5664
- URL: https://www.wjgnet.com/1007-9327/full/v14/i37/5657.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5657
| Age | Sex | Cell type | Stage | Bommann type |
| 51 | M | Moderate | T3N1M0 | II |
| 68 | M | Moderate | T3N1M1 | III |
| 53 | M | Poor with signet ring | T3N3M0 | II |
| 49 | F | Poor | T3N1M0 | II |
| 75 | F | Moderate | T3N1M0 | I |
| 40 | M | Poor | T3N2M0 | III |
| 62 | M | Poor with signet ring | T3N1M0 | II |
| 74 | M | Poor with signet ring | T3N1M0 | II |
| 78 | M | Poor with signet ring | T3N1M0 | III |
| 67 | F | Poor | T3N2M0 | III |
| Spot ID | Protein name | Accession number1 | Top score | Theoretic pI | Theoretic Mr (Da) |
| T1 | Bullous pemphigoid antigen1 isoforms 1/2/3/4/5/8 | Q03001 | 85 | 6.38 | 371 977 |
| T2 | Ras-related protein Rab-30 | Q15771 | 64 | 4.91 | 23 058 |
| T3 | Zinc finger protein 268 | Q14587 | 57 | 9.14 | 108 373 |
| T4 | Calcium-binding protein CaBP5 | Q9NP86 | 61 | 4.46 | 19 812 |
| T5 | RGS3 | P49796 | 62 | 4.79 | 56 566 |
| T61 | MAGUK p55 subfamily member 3 | Q13368 | 45 | 6.27 | 66 168 |
| T7 | Zinc finger protein 134 | P52741 | 57 | 9.18 | 40 297 |
| T81 | Actin, alpha cardiac | P04270 | 154 | 5.23 | 42 019 |
| T9 | Nebulin | P20929 | 59 | 9.10 | 772 727 |
| T10 | Vacuolar protein sorting 33B | Q9H267 | 54 | 6.29 | 70 570 |
| Spot ID | Protein name | Accession number1 | Top score | Theoretic pI | Theoretic Mr (Da) |
| N11 | Peroxiredoxin 2 | P32119 | 72 | 5.66 | 21 892 |
| N21 | Zinc finger protein ZFP-36 | P16415 | 44 | 8.99 | 66 683 |
| N3 | Zinc finger protein 64, isoforms 3 and 4 | Q9NTW7 | 58 | 8.80 | 72 216 |
| N4 | Bullous pemphigoid antigen 1, isoforms 6/9/10 | O94833 | 62 | 5.50 | 590 528 |
| N5 | Transcriptional repressor CTCFL | Q8NI51 | 67 | 8.58 | 75 668 |
Gastric carcinoma is one of the most common malignancies worldwide and is the first most common cause of cancer-related death in China[1]. The main barrier for improving the survival rate is the lack of useful markers for early diagnosis. The cells' fate depends on which gene expresses and how much it expresses. When a cell malignantly transforms, its gene expression profile and their respective protein products may have changed. So it is necessary to isolate and identify genes and proteins that are differentially expressed in gastric cancer.
With the completion of human genome project, the practice of medicine was altered fundamentally, which includes the identification of genes that are involved in the appearance, progression, and treatment of cancer, the answers to what those specific genes do and how they interact in communication networks, and the roles played by their protein products in molecular pathways. However, in most cases, although studies of differential mRNA expression are informative, they do not always correlate with protein concentrations because proteins are often subject to proteolytic cleavage or posttranslational modifications (such as phosphorylation or glycosylation). Cancer biomarkers discovery strategies that target expressed proteins are becoming increasingly popular because proteomic approaches can characterize the proteins, modified or unmodified, involved in cancer progression[2-6].
Recent years, proteomics analysis was has been applied in many kinds of tumors, such as breast cancer[7-9], lung cancer[10,11], prostate cancer[12], liver cancer[13-15] and ovarian cancer[16], etc. In this study, we tried to find biomarkers in human gastric cancer tissue via proteomics as high throughput method. The results presented here will no doubt provide clues to further study of the carcinogenic mechanisms, diagnosis, and therapy of gastric cancer.
Stomach cancer tissues were prepared from ten patients with gastric cancer (the clinical-pathological characteristics are described in Table 1) in Ruijin Hospital. The paired normal gastric tissues were prepared from non cancerous regions at least 5 cm apart from primary tumor. Protean IEF Cell, Protean Plus Multi-Casting Chamber, PROTEAN Plus Dodeca Cell, Fluor-STM Multimager, Immobilized pH gradient (IPG) strips, iodoacetamide, 40% w/v Bio-Lyte pH 3-10 Ampholyte, CHAPS, DTT, urea, acrylamide, N,N′-methylene-bis-acrylamide and RC DC Protein Assay Kit were purchased from Bio-Rad (Richmond, CA, USA). Tris-base, TEMED and SDS were from Promega. Thiourea, DNase, ammonium persulfate and phenylmethyl-sulfonylfluoride (PMSF) were from Sigma-Aldrich (Missouri, USA). 4700 Proteomics Analyzer was from ABI (USA).
Proteins were extracted from human gastric tissue as instructed by the Ha[17] method. Gastric tissue (100 mg) was homogenized in 2 mL buffer (50 mmol/L Tris-HCl, pH 7.2) containing 1 mmol/L PMSF using an homogenizer at 26 000 r/min. The mixture was centrifuged at 1000 g for 5 min to remove debris. Then, the homogenate was centrifuged at 20 000 r/min for 30 min at 4°C. The supernatant was taken as the soluble fraction and TCA (50% w/v) was added to a final concentration of 10% w/v (containing 20 mmol/L DTT) and the solution was allowed to stand on ice for 30 min. The protein precipitate was collected and centrifuged in a microcentrifuge at 14 000 rpm for 10 min at 4°C, and washed three times in 10% TCA (containing 20 mmol/L DTT). The precipitate was washed twice in acetone (containing 20 mmol/L DTT) and dried under an air stream. The dry pellet was dissolved with vortex in the lysis solution (7 mol/L urea, 2 mol/L thiourea, 2% w/v CHAPS, 2% w/v SB3-10, 40 mmol/L Tris, 2 mmol/L TBP, 0.2% w/v Bio-Lyte pH 3-10, 1050 U/mL DNase, 25 μg/mL RNase) and allowed to stand for 1 h at room temperature. After centrifugation at 14 000 r/min for 10 min at 15°C, the supernatant was used as the two-dimensional polyacrylamide gel electrophoresis (2-DE) sample for the soluble fraction. The protein samples were stored in aliquots at -80°C until use. Protein concentration of 2-DE samples was determined using RC DC Protein Assay kit.
The first-dimensional gel separation was carried out with 17 cm IPG strips (pH 3-10, pH 4-7, pH 5-8) following the manufacturers’ protocol with minor modifications. Samples containing up to 100 μg protein were diluted to 320 μL with rehydration solution (7 mol/L urea, 2 mol/L thiourea, 2% CHAPS, 100 mmol/L DTT, 0.2% w/v Bio-Lyte pH 3-10, trace bromophenol blue), and applied to strips by overnight rehydration at 50 V. Proteins were focused succeedingly for 1 h at 150 V, 1 h at 250 V, 1 h at 1000 V, then a gradient was applied from 1000 to 10 000 V in 5 h, and focusing was continued at 10 000 V for 6 h to give a total of 88 kVh on a Protean IEF Cell. After IEF, strips were equilibrated for 15 min in 6 mol/L urea, 2% SDS, 0.05 mol/L Tris-HCl, pH 8.8, and 30% glycerol containing 2% DTT, and then equilibrated again for 15 min in the same buffer containing 2.5% iodoacetamide. Equilibrated IPG strips were transferred onto 12% uniform polyacrylamide gels and then were run in Protean Plus Dodeca Cell (Bio-Rad, USA) at 24 mA per gel for 5 h and 30 min. The gels were visualized using silver staining method. After staining, 2D gels were scanned using Fluor-STM Multimager (Bio-Rad, USA) and images were analyzed using PDQuest 7.1.1 (Bio-Rad, USA).
Protein spots of interest were cut manually from the gel and placed into siliconized microcentrifuge tubes. The gel fragments were destained in solution that consisted of 100 mmol/L Na2S2O3 and 30 mmol/L K3Fe(CN)6 (V/V, 1:1) and dehydrated by washing two times for 10-15 min with 25 mmol/L NH4HCO3 in 50% acetonitrile until shrunken and white. The protein-containing gel-spots were reduced in the reduction buffer (25 mmol/L NH4HCO3 and 10 mmol/L DTT) for 1 h at 56°C and then alkylated in the alkylation buffer (25 mmol/L NH4HCO3 and 55 mmol/L iodoacetamide) in the dark for 30 min at room temperature. The gel pieces were washed twice with 100 mmol/L NH4HCO3 and dehydrated by 100% acetonitrile. Ten microliters modified trypsin (8 μg/mL) was added to each tube. Tubes were sealed with parafilm and digestion was performed for 16 h at 30°C. When digestion was completed, 100 μL of 100 mmol/L NH4HCO3 was added and tubes were sonicated for 10 min. The supernatant was removed from each tube and transferred into a fresh tube. Further recovery of the peptides was accomplished by two extractions with 50% acetonitrile/5% TFA. All supernatants were pooled and concentrated and desalted by ZIPTIPS (Millipore, USA) prior to applying on to the sample plate.
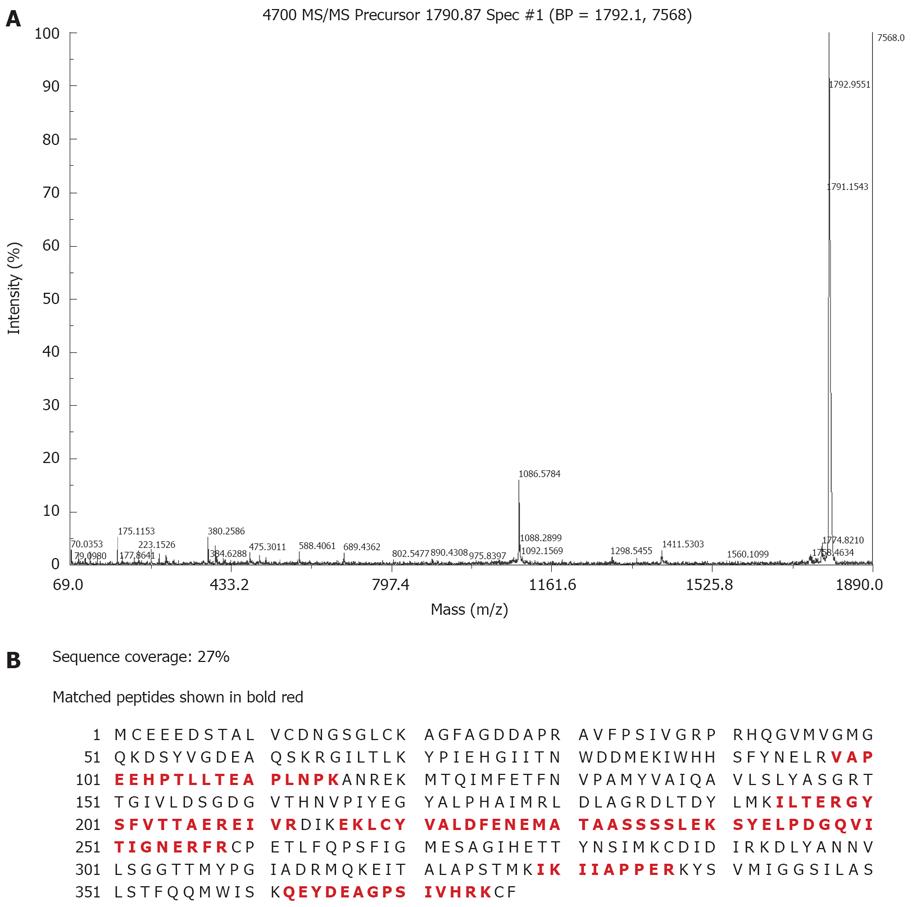
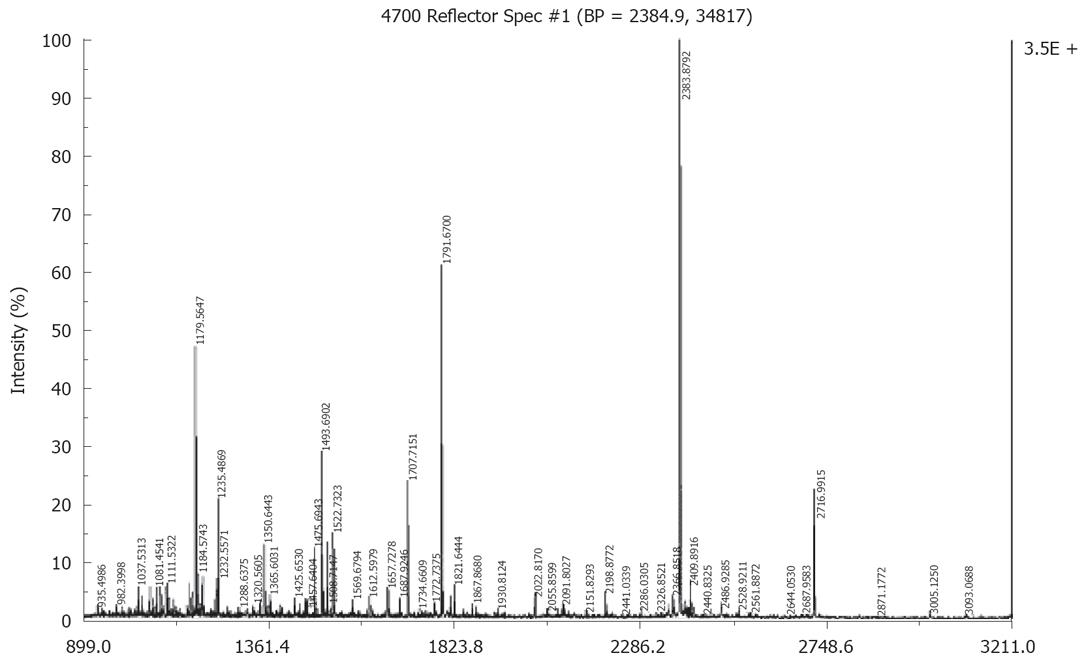
A small fraction (0.5 μL) of the sample was mixed with a-cyano-4-hydroxycinnamic acid matrix (1:1, v/v) and the mixture solution was then spotted on the plate and dried, then analyzed on a 4700 MALDI-TOF-TOF mass spectrometer (Applied Biosystems, USA). The laser wavelength was 355 nm, and the laser repetition rate was 200 Hz. External calibration was employed. Samples that did not get positive identification by MALDI were subjected to MALDI-TOF-TOF-MS using a 4700 Proteomics Analyzer equipped with a special analysis software GPS and local databases. The following parameters were used in the searches: SWISS-PROT was used as the protein sequence database; trypsin digest (one missed cleavage allowed); mass accuracy 0.2 Da; species: Homo sapiens; acetylation of the N-terminus, alkylation of cysteine by carbamidomethylation and oxidation of methionine were considered as possible modifications.
Resolution and reproducibility of 2-DE have been improved significantly since the introduction of IPG strips[18,19]. However, it is still a time-consuming and laborious job. In this study, we firstly separate the soluble proteins of human gastric tissue in IPG strips with different pH intervals. All maps (Figure 1) were constructed on silver-stained gels with protein loadings of 100 μg. Proteins from normal and tumor tissues of stomach were separated in pH 3-10 IPG strips. After SDS-PAGE, about 850 protein spots could be detected (Figure 1A and B). While, about 670 protein spots could be detected which separated by pH 4-7 IPG strips (Figure 1C and D). The well-resolved, reproducible 2-DE patterns of human gastric cancer tissues and paired normal tissues were obtained with pH 5-8 IPG strips (Figure 1E and F). About 1400 spots were detected, respectively, and about a total of 1100 spots were matched each other. We got better resolution and more protein spots with pH 5-8 IPG strips; therefore, in the succeeding studies, we chose pH 5-8 IPG strips to analyze proteins of human gastric tissues.
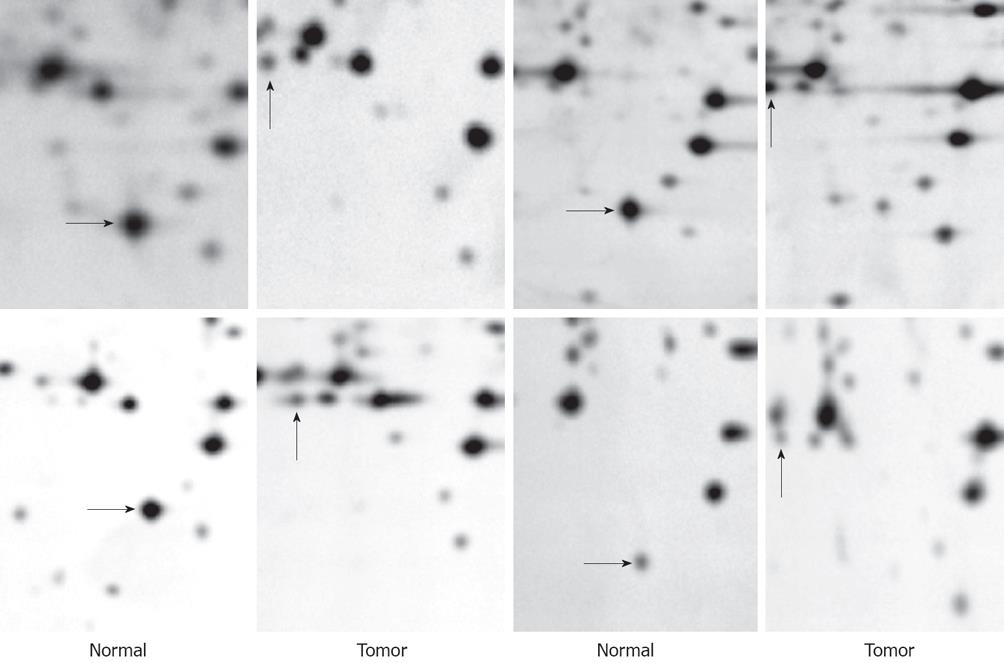
After 2-DE, the differential proteins between human gastric cancer tissues and paired normal tissues were found by PDQuest 7.1.1 2-D gel analysis software (BIO-RAD, USA). To compare 2-DE maps, it is important to have a representative sample. Hence, an average electrophoresis map of human gastric cancer tissues was constructed by the combination of the 2-DE maps from 10 tumor tissues with the PDQuest 7.1.1 2-DE gel analysis software. Similarly, an average electrophoresis map of 10 normal gastric tissues was also established. These average electrophoresis maps were used to perform the differential expression analysis. The differential protein spots with at least five-fold discrepancy, which were present in at least four paired tumor and normal tissues, were detected by comparing the 2-DE protein patterns of the average gels of tumor and control tissues. Figure 2 shows differentially expressed proteins of N4, T8 between tumor and control tissues in four patients. Twenty three protein spots were selected to be identified by mass spectrometry (MS). Among these, 16 spots were over expressed and other 7 spots were under expressed in gastric cancer tissue.
Fifteen proteins out of the selected 23 proteins were identified successfully by MALDI-TOF MS or tandem MS (Figures 3 and 4) and there were ten over expressed and five under expressed proteins in stomach cancer tissues compared with normal tissues. The identified proteins are shown in Tables 2 and 3.
Proteins which were up-regulated in gastric cancer tissue includes: (1) transcriptional regulation proteins, such as zinc finger protein; (2) signal transduction regulator, such as regulator of G-protein signaling 3 (RGS3); (3) cancer-related proteins, such as ras-related protein; (4) cytoskeleton proteins, such as bullous pemphigoid antigen 1 isoforms 1/2/3/4/5/8; (5) tissue-specific proteins, such as Calcium-binding protein CaBP5. Proteins which were down-regulated in gastric cancer tissue includes: (1) transcriptional regulation proteins, such as zinc finger protein and transcriptional repressor CTCFL; (2) cytoskeleton proteins, such as bullous pemphigoid antigen 1 isoforms 6/9/10; (3) metabolic enzymes, such as peroxiredoxin 2.
Genomics-based techniques, such as expressed sequence tag (EST) sequencing, serial analysis of gene expression (SAGE)[20], cDNA expression array hybridization[21], were widely used to find out potential cancer biomarkers. However, a major limitation to these approaches is detection of high levels of mRNA transcripts of a specific gene may not necessarily mean that high levels of the corresponding protein product will be present. Now, proteomics-based techniques can help us find cancer-related biomarkers at the protein level.
In this study, we obtained ten over expressed proteins (Bullous pemphigoid antigen 1 isoforms 1/2/3/4/5/8, Ras-related protein Rab-30, Zinc finger protein 268, Calcium-binding protein CaBP5, RGS3, membrane-associated guanylate kinase (MAGUK) p55 subfamily member 3, Zinc finger protein 134, Actin alpha cardiac, Nebulin, Vacuolar protein sorting 33B) in gastric cancer tissue.
RGS3 inhibits signal transduction by increasing the GTPase activity of G protein alpha subunits thereby driving them into their inactive GDP. A recent study revealed that RGS3 gene was up-regulated in highly migratory clones selected from U373MG glioma cells compared with that of the original cells using oligonucleotide microarrays comprising 12 625 genes. U373MG glioma cell clones with over-expression of RGS3 or RGS4 showed an increase of both adhesion and migration. These findings expand the spectrum of possible molecular pathways underlying the invasion of neoplastic astrocytes[22].
MAGUK p55 subfamily member 3 (MPP3) belongs to the MAGUK protein family. Shins' study showed MPP3 was identified by cell surface biotinylation and MS in the cell surface proteome of A549 lung adenocarcinoma cells and LoVo colon adenocarcinoma cells[23].
Nebulin may be involved in maintaining the structural integrity of sarcomeres and the membrane system associated with the myofibrils. The nebulin gene was found up-regulated in non small cell lung carcinoma-derived cell lines D51 and lung cancer cell lines (H2170 and H526) compared with human bronchial epithelial cell lines by suppression subtractive hybridization (SSH). But the relations between nebulin and gastric cancer is not clear[24].
We also obtained five under-expressed proteins (Peroxiredoxin 2, Zinc finger protein ZFP-36, Zinc finger protein 64 isoforms 3 and 4, Bullous pemphigoid antigen 1 isoforms 6/9/10, Transcriptional repressor CTCFL).
Peroxiredoxins (Prxs) are a novel group of peroxidases containing high antioxidant efficiency and some of them having also effects on cell differentiation and apoptosis. The mammalian Prx family has six distinct members located in various subcellular locations. Peroxiredoxin 2 (Prx 2) is known not only to protect cells from oxidative damage caused by hydrogen peroxide (H2O2), but also to endow cancer cells with resistance to both H2O2 and cisplatin and to grant them radioresistance[25]. Somiari et al[26] analyzed human breast infiltrating ductal carcinoma (IDCA) using two-dimensional difference gel electrophoresis and MS and found peroxiredoxin 2 was less abundant in four breast IDCAs (StageI, IIA, IIB and IIIA) compared with non-neoplastic tissue. But recent studies showed Prx family were over expressed in some kind of carcinomas suggesting that Prx has a proliferative effect and may be related to cancer development or progression[27-30].
The transcriptional repressor, CTCFL, acts as a transcriptional regulator binding to some gene promoters and may be associated with epigenetic reprogramming events in the male germ line. But the role that transcriptional repressor CTCFL plays in gastric cancer is still not demonstrated.
Because of different protein processing and posttranslational modifications, protein isoforms of individual protein can be identified on 2D gels. Interestingly, our study revealed Bullous pemphigoid antigen 1 isoforms 1/2/3/4/5/8 were over expressed in gastric cancer tissue while Bullous pemphigoid antigen 1 isoforms 6/9/10 were under expressed in cancer tissue. This finding suggests that the different protein isoforms may be correlated with specific features of gastric cancer and play different roles in the progression of cancer.
In conclusion, the identified proteins, both up-regulated and down-regulated, may participate in the progression of malignant growth or in the maintenance of normal conditions of gastric tissue. The functions of these proteins in the carcinogenesis of gastric cancer remain to be studied. We hope they may provide useful information for early detection and therapy of gastric cancer.
Gastric carcinoma is one of the most common malignancies worldwide and is the most common cause of cancer-related death in China. The main barrier for improving survival rate is the lack of useful markers for early diagnosis.
Identification differential proteins between tumor tissue and normal tissue by proteomics to be candidate tumor markers for gastric cancer.
Twenty three differential proteins were found between tumor and normal tissues of gastric cancer, among these fifteen proteins were identified.
The differential proteins may be tumor markers for gastric cancer.
The authors utilized two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to isolate and identify differentially expressed proteins between gastric cancer tissues and paired normal mucosa. This is an interesting paper using new technology to identify gastric cancer characteristics.
Peer reviewer: Yaron Niv, Professor, Department of Gastroenterology, Rabin Medical Center, Beilinson Campus, Tel Aviv University, 2 Hadekel St., Pardesia 42815, Israel
S- Editor Li DL L- Editor Rippe RA E- Editor Ma WH
| 1. | Sun XD, Mu R, Zhou YS, Dai XD, Zhang SW, Huangfu XM, Sun J, Li LD, Lu FZ, Qiao YL. [Analysis of mortality rate of stomach cancer and its trend in twenty years in China]. Zhonghua Zhongliu Zazhi. 2004;26:4-9. |
| 3. | Simpson RJ, Bernhard OK, Greening DW, Moritz RL. Proteomics-driven cancer biomarker discovery: looking to the future. Curr Opin Chem Biol. 2008;12:72-77. |
| 4. | Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267-275. |
| 5. | Bitarte N, Bandres E, Zarate R, Ramirez N, Garcia-Foncillas J. Moving forward in colorectal cancer research, what proteomics has to tell. World J Gastroenterol. 2007;13:5813-5821. |
| 6. | Cowan ML, Vera J. Proteomics: advances in biomarker discovery. Expert Rev Proteomics. 2008;5:21-23. |
| 7. | Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296-1304. |
| 8. | Clarke CH, Buckley JA, Fung ET. SELDI-TOF-MS proteomics of breast cancer. Clin Chem Lab Med. 2005;43:1314-1320. |
| 9. | Rui Z, Jian-Guo J, Yuan-Peng T, Hai P, Bing-Gen R. Use of serological proteomic methods to find biomarkers associated with breast cancer. Proteomics. 2003;3:433-439. |
| 10. | Li C, Chen Z, Xiao Z, Wu X, Zhan X, Zhang X, Li M, Li J, Feng X, Liang S. Comparative proteomics analysis of human lung squamous carcinoma. Biochem Biophys Res Commun. 2003;309:253-260. |
| 11. | Au JS, Cho WC, Yip TT, Law SC. Proteomic approach to biomarker discovery in cancer tissue from lung adenocarcinoma among nonsmoking Chinese women in Hong Kong. Cancer Invest. 2008;26:128-135. |
| 12. | Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu Q, Xu GJ, Song JD, Zhao FK. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121:2596-2605. |
| 13. | Kim J, Kim SH, Lee SU, Ha GH, Kang DG, Ha NY, Ahn JS, Cho HY, Kang SJ, Lee YJ. Proteome analysis of human liver tumor tissue by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of disease-related proteins. Electrophoresis. 2002;23:4142-4156. |
| 14. | Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470-477. |
| 15. | Feng Y, Tian ZM, Wan MX, Zheng ZB. Protein profile of human hepatocarcinoma cell line SMMC-7721: identification and functional analysis. World J Gastroenterol. 2007;13:2608-2614. |
| 16. | Gagnon A, Kim JH, Schorge JO, Ye B, Liu B, Hasselblatt K, Welch WR, Bandera CA, Mok SC. Use of a combination of approaches to identify and validate relevant tumor-associated antigens and their corresponding autoantibodies in ovarian cancer patients. Clin Cancer Res. 2008;14:764-771. |
| 17. | Ha GH, Lee SU, Kang DG, Ha NY, Kim SH, Kim J, Bae JM, Kim JW, Lee CW. Proteome analysis of human stomach tissue: separation of soluble proteins by two-dimensional polyacrylamide gel electrophoresis and identification by mass spectrometry. Electrophoresis. 2002;23:2513-2524. |
| 18. | Bjellqvist B, Ek K, Righetti PG, Gianazza E, Gorg A, Westermeier R, Postel W. Isoelectric focusing in immobilized pH gradients: principle, methodology and some applications. J Biochem Biophys Methods. 1982;6:317-339. |
| 19. | Gorg A, Postel W, Gunther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531-546. |
| 20. | Yasui W, Oue N, Ito R, Kuraoka K, Nakayama H. Search for new biomarkers of gastric cancer through serial analysis of gene expression and its clinical implications. Cancer Sci. 2004;95:385-392. |
| 21. | Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397-414. |
| 22. | Tatenhorst L, Senner V, Puttmann S, Paulus W. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J Neuropathol Exp Neurol. 2004;63:210-222. |
| 23. | Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, Zhao R, Puravs E, Tra J, Michael CW. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607-7616. |
| 24. | Difilippantonio S, Chen Y, Pietas A, Schluns K, Pacyna-Gengelbach M, Deutschmann N, Padilla-Nash HM, Ried T, Petersen I. Gene expression profiles in human non-small and small-cell lung cancers. Eur J Cancer. 2003;39:1936-1947. |
| 25. | Yo YD, Chung YM, Park JK, Ahn CM, Kim SK, Kim HJ. Synergistic effect of peroxiredoxin II antisense on cisplatin-induced cell death. Exp Mol Med. 2002;34:273-277. |
| 26. | Somiari RI, Sullivan A, Russell S, Somiari S, Hu H, Jordan R, George A, Katenhusen R, Buchowiecka A, Arciero C. High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast. Proteomics. 2003;3:1863-1873. |
| 27. | Kinnula VL, Paakko P, Soini Y. Antioxidant enzymes and redox regulating thiol proteins in malignancies of human lung. FEBS Lett. 2004;569:1-6. |
| 28. | Karihtala P, Mantyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9:3418-3424. |
| 29. | Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21:2085-2090. |
| 30. | Yanagawa T, Iwasa S, Ishii T, Tabuchi K, Yusa H, Onizawa K, Omura K, Harada H, Suzuki H, Yoshida H. Peroxiredoxin I expression in oral cancer: a potential new tumor marker. Cancer Lett. 2000;156:27-35. |