Published online Dec 21, 2007. doi: 10.3748/wjg.v13.i47.6370
Revised: August 26, 2007
Accepted: November 19, 2007
Published online: December 21, 2007
AIM: To investigate the change in eukaryotic gene expression profile in Caco-2 cells after infection with strains of Escherichia coli and commensal probiotic bacteria.
METHODS: A 19200 gene/expressed sequence tag gene chip was used to examine expression of genes after infection of Caco-2 cells with strains of normal flora E. coli, Lactobacillus plantarum, and a combination of the two.
RESULTS: The cDNA microarray revealed up-regulation of 155 and down-regulation of 177 genes by E. coli. L. plantarum up-regulated 45 and down-regulated 36 genes. During mixed infection, 27 genes were up-regulated and 59 were down-regulated, with nullification of stimulatory/inhibitory effects on most of the genes. Expression of several new genes was noted in this group.
CONCLUSION: The commensal bacterial strains used in this study induced the expression of a large number of genes in colonocyte-like cultured cells and changed the expression of several genes involved in important cellular processes such as regulation of transcription, protein biosynthesis, metabolism, cell adhesion, ubiquitination, and apoptosis. Such changes induced by the presence of probiotic bacteria may shape the physiologic and pathologic responses they trigger in the host.
-
Citation: Panigrahi P, Braileanu GT, Chen H, Stine OC. Probiotic bacteria change
Escherichia coli -induced gene expression in cultured colonocytes: Implications in intestinal pathophysiology. World J Gastroenterol 2007; 13(47): 6370-6378 - URL: https://www.wjgnet.com/1007-9327/full/v13/i47/6370.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i47.6370
There has been an upsurge in clinical trials involving probiotics in gastrointestinal diseases. Although promising, these trials with specific probiotic bacteria have shown variable results, with limited elucidation of the underlying pathophysiology. In real life, these strains never act on the host cells in isolation and over 800 bacterial strains in the adult human colon are engaged in constant cross talk with intestinal epithelial cells. No detailed study so far has attempted to examine the effect of individual probiotic bacteria on host gastrointestinal cells, and the changes during co-infection with other enteric bacteria.
However, a lot of emphasis has recently been given to the normal bacterial flora in the intestine, including many Lactobacillus strains that are considered as probiotics with health-promoting effects on the host. A myriad of effects have been shown by these bacteria, spanning from bacterial killing via secretion of bacteriocins[1], to inhibition of attachment and invasion by pathogenic bacteria[2], and modulation of host inflammatory responses[3]. These commensal strains have been shown to modulate the expression of genes involved in important physiologic functions such as postnatal intestinal maturation, cell growth, proliferation, nutrient absorption, mucosal barrier function, and angiogenesis[4-6]. Multiple laboratory studies have shown beneficial effects of Lactobacillus strains against single pathogenic bacterial strains in in vitro and in vivo systems[7,8]. During the last few years, there has been an exponential increase of clinical trial reports and reviews in the literature pertaining to the utility of probiotics in gastrointestinal and allergic diseases[9-21]. Many small studies utilizing Lactobacillus and Bifidobacteria have shown beneficial effects such as better weight gain and improved feeding tolerance[22] in neonates, and efficacy against neonatal necrotizing enterocolitis (NEC)[23-25] and sepsis[24]. Other reports have demonstrated no effect in NEC[26], and in some cases, deterioration of specific conditions with probiotic therapy[25]. Results of clinical trials done by our group have shown a wide range (0%-60%) of colonization rates in newborn infants when three different probiotic strains were used[27]. These mixed and non-reproducible results have raised more questions than providing answers, and have strongly suggested complex interactions among bacterial strains and epithelial cells in the human intestine[7,28-30].
At this time, our understanding of the response of eukaryotic cells (e.g., intestinal cells) is limited to nutrients and local factors[31], and virulence mechanisms involving individual microorganisms. Although contrasting signal transduction mechanisms in bacterial and eukaryotic gene transcription have been described[32], reports on cross talk between bacteria and epithelial cells have focused on single bacterial strains[33]. As a result, the physiologic and pathologic changes in the host cells as a response to multiple bacteria have not been addressed. Since the mammalian gut is colonized with multiple bacterial strains very quickly after birth, it is conceivable that the ultimate effect of probiotic treatment will depend greatly on the presence of other bacteria in the host intestine at that time.
In the current study, we examined the difference between gene expression in intestinal cells in response to infection with a single bacterial strain, compared to that during mixed infection. Caco-2 cells were utilized to discern the effect of Lactobacillus plantarum (the most common Lactobacillus species in humans)[34], Escherichia coli (a common Gram-negative enteric strain) and the combination of the two strains. A high-density cDNA glass microarray and standard techniques were employed to identify bacteria-induced gene expression in this eukaryotic system.
Caco-2 cells, obtained from American Type Culture Collection (ATCC HTB-37), were used at passage 10-12. This human colon-adenocarcinoma-derived cell line has been used extensively for physiologic and enteric bacterial pathogenesis studies[35]. The cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA with 10% fetal calf serum (Sigma, St. Louis, MO, USA), 2 mmol/L glutamine, 1.0 mmol/L sodium pyruvate, 0.1% non-essential amino acids, 100 U/mL penicillin and 100 μg/mL streptomycin. All experiments were performed without serum or antibiotics in 8-10-d-old cells after they reached confluence.
E. coli strain 6-1 was isolated from a healthy infant, and has been used previously in in vitro and in vivo studies in our laboratory[36]. This strain does not possess any known virulence genes[37]. We used a human strain of L. plantarum (ATCC 202195), the species most commonly isolated from humans[34].
Cells were washed in PBS and re-fed with experimental DMEM without serum or antibiotics before the experiments. Following previously described methods in which a maximal effect of Lactobacillus was seen, Caco-2 cells were infected with E. coli and/or L. plantarum at 1:10 multiplicity of infection, and incubated for 2 h[38].
For examination of Caco-2 cell gene expression under our experimental conditions, we used a high-density glass microarray H19K (University Health Network Microarray Centre, Toronto, http://www.microarrays.ca/home.html) that had 19200 genes/expressed sequence tags (ESTs). These included fully characterized, partially characterized and some uncharacterized human gene elements. Each gene/EST was printed in duplicate in this array. The genes in the array represented constitutively expressed genes/ESTs and the manufacturer did not include genes that are transiently expressed, such as cytokines and chemokines. In our experiments, we used dye swapping procedures and bioinformatics tools considered as standard techniques that have been reported in similar studies in the past[39].
Total RNA was extracted from Caco-2 cells grown in 75-cm2 tissue culture flasks using the TRIZOL method (Invitrogen, Carlsbad, CA, USA), following manufacturer’s instructions. RNA samples were treated with RNAse-free DNAse to remove contaminating genomic DNA, examined by 260/280 nm UV absorption ratio (> 1.8) followed by assessment of integrity by running in a 1.2% agarose gel and ethidium bromide staining.
Total mRNA (10 μg) was reversely transcribed using 20 mmol/L dNTP mix including amino-allyl dUTP (AA-dUTP; Sigma) and 400 U SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). The resulting aa-cDNA, cleaned with a QIAquick column (Qiagen, Valencia, CA, USA), was coupled with Cy3 or Cy5 dye (Amersham Biosciences, Piscataway, NJ, USA) in the presence of sodium bicarbonate for 1 h in the dark. After adding 10 μL 4 mol/L hydroxylamine and 125 μL buffer PB (Qiagen supplied) to each, the control and treatment samples were combined and cleaned using another QIAquick column. The elute was transferred to a Microcon YM 30 centrifugal filter device (Amicon Millipore, Bedford, MA, USA), and after adding 20 μL cot-1 human DNA (Gibco-BRL), the whole volume was concentrated to 5 μL. Ten microliters of 1 μg/μL poly (A) RNA (Sigma), 1 μL 10 μg/μL tRNA (Gibco-BRL), 4 μL water and 5 μL hybridization buffer (50% formamide, 5 × SSC (3 mol/L sodium chloride, 0.3 mol/L sodium citrate and 0.1% SDS) were added. The array was pretreated at 42°C for 1 h with hybridization buffer. After overnight hybridization at 42°C, the slides were washed in 50 mL 2 × SSC and 0.1% SDS at 55°C for 5 min, once in 0.1 × SSC and 0.1% SDS for 5 min at room temperature (RT), and for 5 min with 0.1 × SSC at RT, air-dried and scanned with 555 nm and 647 nm lasers in a Scan Array 5000 (GSI Lumonics, Novi, MI, USA). Images of the fluorescence intensity for each dye were analyzed using Imagene 4.2 software (Biodiscovery, CA, USA).
RNA from each experimental condition and control Caco-2 cells were hybridized on the same microarray. To eliminate the color bias, duplicate reactions were carried out in which the dyes (Cy3, Cy5) for the control and experimental samples were swapped.
Individual gene intensity data files for each experimental condition were compared with the control values using the GeneSight 2.1 program (Biodiscovery). After correction for the local background, normalization using all the spots, removal of the outliers, averaging of the replicates and transforming to base 2, each gene was assigned a relative expression value when compared with the control. A twofold or larger difference in the relative gene expression was considered significant.
The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE5874.
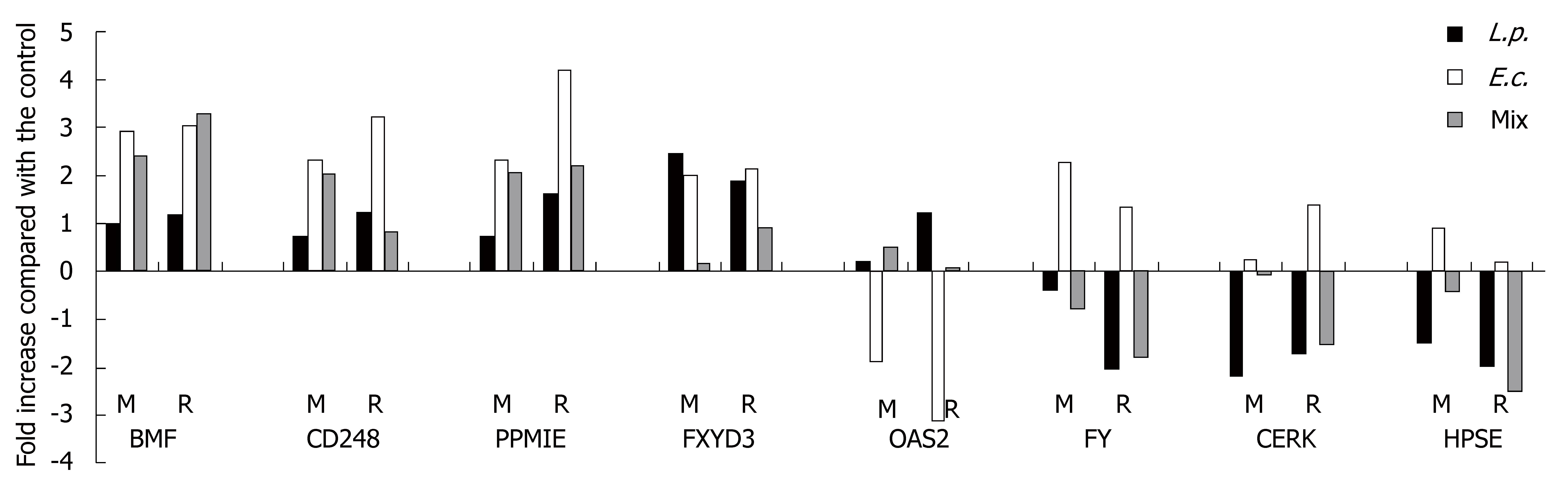
We randomly selected eight genes (BMF, CD248, PPM1E, FXYD3, OAS2, FY, CERK and HPSE) from our pool of expressed genes/ESTs that are well characterized in the literature and appear to have some biologic significance. ESTs were not included. Real-time quantitative PCR (Bio-Rad iQ SYBR Green Supermix and iCicler) was done using GAPDH for normalization. The levels of expression detected by microarray were compared with PCR results. The primers used to amplify specific gene segments are presented in Table 1. The relative gene expression was calculated using the comparative ΔΔCT method. Each sample was tested twice in triplicate.
| Nr. | Gene symbol | Gene ID NCBI | Gene name | Location | Function | Relative fold modification | ||
| L.p. | E.c. | Mix | ||||||
| 1 | GPR34 | 2857 | G protein-coupled receptor 34 | Integral to plasma membrane | G-protein coupled receptor activity | 2.43 | 3.03 | -0.53 |
| 2 | GTPBP4 | 23560 | GTP binding protein 4 | Nucleus | Ribosome biogenesis - small GTPase mediated signal transduction | 2.00 | 2.91 | -0.30 |
| 3 | TFPI2 | 7980 | Tissue factor pathway inhibitor 2 | Extracellular matrix | Serine-type endopeptidase inhibitor activity | 2.10 | 2.93 | -0.27 |
| 4 | CYP26A1 | 1592 | Cytochrome P450, family 26,subfamily A, polypeptide 1 | Membrane | Metal ion binding | 2.31 | 2.39 | -0.88 |
| 5 | ZNF35 | 7584 | Zinc finger protein 35 (clone HF.10) | Nucleus | Transcription factor activity | 2.18 | 2.19 | -0.66 |
| 6 | RTTN | 25914 | Rotatin | required for left-right specification in mouse embryos | 2.21 | 2.13 | -0.58 | |
| 7 | FXYD3 | 5349 | FXYD domain containing ion transport regulator 3 | Membrane | Chloride channel activity | 2.44 | 2.03 | -0.15 |
| 8 | CYYR1 | 116159 | Cysteine/tyrosine-rich 1 | Integral to membrane | Molecular function unknown | -2.30 | -2.20 | 1.05 |
| 9 | BFAR | 512836 | Bifunctional apoptosis regulator | Apoptosis regulator | -2.40 | -2.17 | 1.06 | |
| 10 | C19orf4 | 25789 | Chromosome 19 open reading frame 4 | Integral to, membrane | Molecular function unknown | -2.51 | -2.28 | 0.19 |
| 11 | KIAA1305 | 57523 | KIAA1305 protein | Hypothetical protein | -2.19 | -2.29 | 0.55 | |
| 12 | PCDH9 | 5101 | Protocadherin 9 | Integral to membrane | Cell adhesion | -2.18 | -2.30 | 0.50 |
| 13 | IKIP | 121457 | IKK interacting protein | -2.23 | -2.33 | 0.83 | ||
| 14 | FLJ21963 | 79611 | FLJ21963 protein | Hypothetical protein | -2.12 | -2.37 | 1.88 | |
| 15 | SCRG1 | 11341 | Scrapie responsive protein 1 | Extracellular space | Nervous system development | -2.14 | -2.54 | 0.74 |
| 16 | ULK2 | 9706 | Unc-51-like kinase 2 (C. elegans) | Similar to a serine/threonine kinase in C. elegans | -2.46 | -2.72 | 0.26 | |
| 17 | LIFR | 3977 | Leukemia inhibitory factor receptor | Integral to plasma membrane | Receptor activity | -2.26 | -2.96 | 0.29 |
| 18 | BMF | 90427 | Bcl-2 modifying factor | Sequestrated by myosin | Apoptotic activator - protein binding | 1.00 | 2.95 | 2.40 |
| 19 | CD248 | 57124 | CD248 antigen, endosialin | Marker of stromal fibroblasts | 0.74 | 2.31 | 2.10 | |
| 20 | PPM1E | 22843 | Protein phosphatase 1E (PP2C domain containing) | Phosphatase | 0.76 | 2.33 | 2.06 | |
| 21 | CARD8 | 22900 | Caspase recruitment domain family, member 8 | Involved in NFκB pathway | -0.59 | -3.26 | 4.07 | |
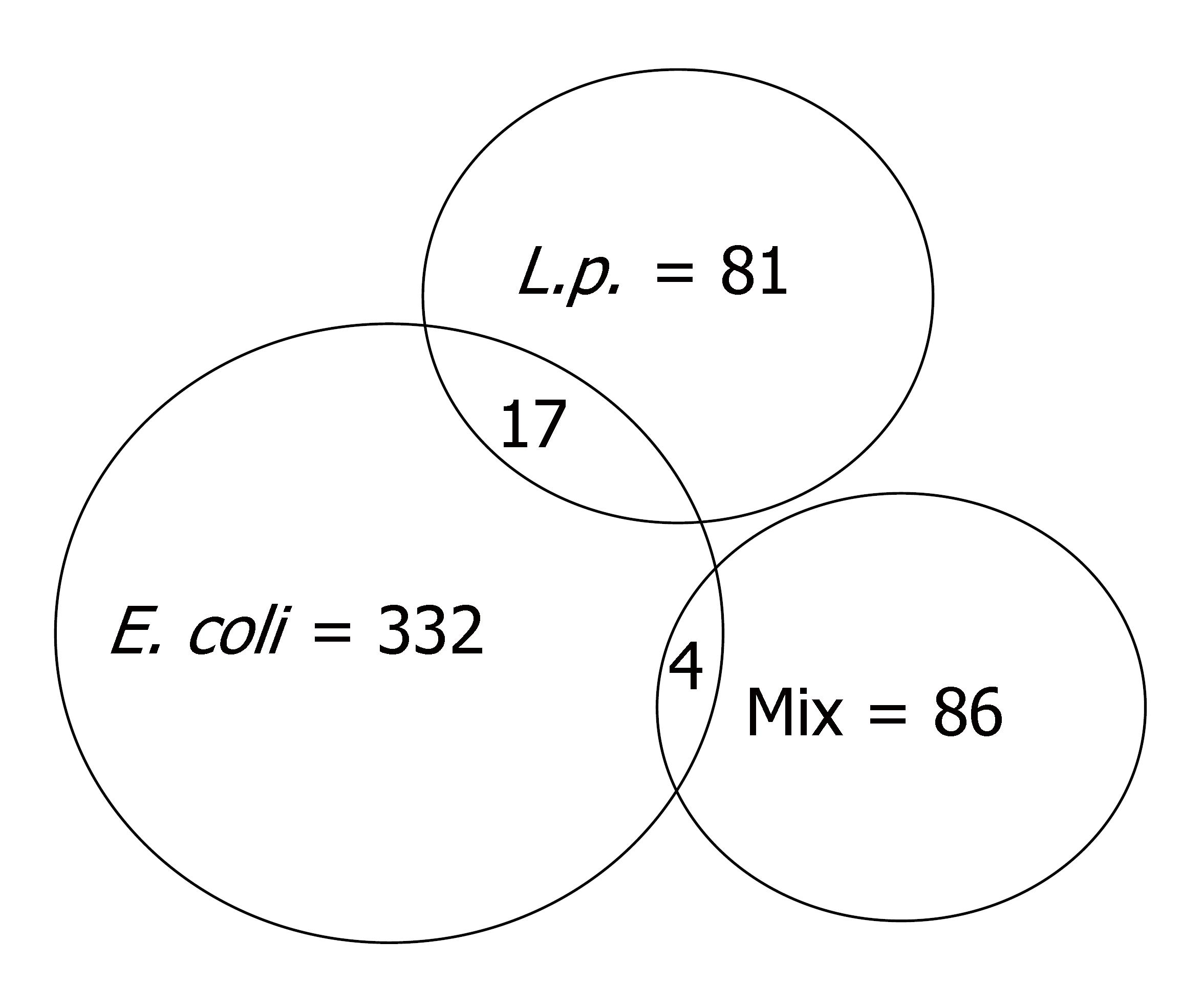
After 2 h treatment, E. coli, L. plantarum and their combination changed the expression (by twofold) of 332, 81 and 86 genes, respectively, compared to uninfected control Caco-2 cells (Figure 1). After infection with E. coli, 155 genes were up-regulated and 177 were down-regulated (Table 1 and Supplementary Table 1). L. plantarum induced up-regulation of 45 genes and 36 genes were down-regulated (Table 1 and Supplementary Table 2). The combination treatment up-regulated 27 genes and down-regulated 59 (Table 1 and Supplementary Table 3) [Note: The supplementary tables above can be accessed at: http://panigrahipeds.googlepages.com/suppl-tables.pdf; Raw data of all 19200 genes during each treatment can be accessed from the NCBI/GEO data base (GSE5874) at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=nzyxdkkuwukuytk&acc=GSE5874]. Mixed infection nullified the previously demonstrated stimulatory and inhibitory effects of E. coli on 152 and 177 genes and of L. plantarum on 38 and 26 genes, respectively. Stimulation of 23 and inhibition of 59 genes were noted after mixed infection that was not influenced by either bacterium alone.
| Biological process | Gene symbol | Gene ID NCBI | Gene name | Fold change |
| Category 1: Regulation of transcription | HOXD10 | 3236 | Homeobox D10 | 2.50 |
| PHF7 | 51533 | PHD finger protein 7 (Zinc ion binding) | 2.44 | |
| EGR1 | 1958 | Early growth response 1 | -2.08 | |
| TRIM24 | 8805 | Tripartite motif-containing 24 (Zinc ion and DNA binding) | -2.15 | |
| ENO1 | 2023 | Enolase 1, (alpha) (DNA binding) | -2.34 | |
| Category 2: RNA processing | SSB | 6741 | Sjogren syndrome antigen B (autoantigen La) | -2.08 |
| FUSIP1 | 10772 | FUS interacting protein (serine/arginine-rich) 1 | -2.21 | |
| NOLA5 | 10528 | Nucleolar protein 5A (56 kDa with KKE/D repeat) | -2.27 | |
| DDX5 | 1655 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | -2.71 | |
| Category 3: Protein biosynthesis, folding, binding and transport | NEURL | 9148 | Neuralized homolog (Intracellular protein transport) | 2.91 |
| WDR36 | 134430 | WD repeat domain 36 | 2.37 | |
| MTFMT | 123263 | Mitochondrial methionyl-tRNA formyltransferase | 2.04 | |
| ARF4 | 378 | ADP-ribosylation factor 4 | -2.03 | |
| CEP57 | 9702 | Centrosomal protein 57 kDa | -2.11 | |
| ETF1 | 2107 | Eukaryotic translation termination factor 1 | -2.27 | |
| HSPA1A | 3303 | Heat shock 70 kDa protein 1A | -2.28 | |
| LGALS3 | 3958 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | -2.75 | |
| HSPH1 | 10808 | Heat shock 105 kDa/110 kDa protein 1 | -2.93 | |
| HSPA8 | 3312 | Heat shock 70 kDa protein 8 | -2.97 | |
| Category 4: Structural protein | AMPH | 273 | Amphiphysin (Actin cytoskeleton) | 3.04 |
| MAP1B | 4131 | Microtubule-associated protein 1B | 2.13 | |
| ABCC10 | 89845 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | -2.05 | |
| SLC26A2 | 1836 | Solute carrier family 26 (sulfate transporter), member 2 | -2.06 | |
| TUBB2A | 7280 | Tubulin, beta 2A | -2.15 | |
| Category 5: Metabolism | C5orf14 | 79770 | Chromosome 5 open reading frame 14 | 2.91 |
| NAV2 | 89797 | Neuron navigator 2 | 2.83 | |
| SLC24A4 | 123041 | Solute carrier family 24 (sodium/potassium/calcium), member 4 | 2.35 | |
| PLEKHM2 | 23207 | Pleckstrin homology domain containing, family M, member 2 | 2.10 | |
| TWF1 | 5756 | Twinfilin, actin-binding protein, homolog 1 (Tyrosin kinase) | -2.01 | |
| AKR1C1 | 1645 | Aldo-keto reductase 1, member C1 (Bile acid binding) | -2.09 | |
| HMGCR | 3156 | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | -2.10 | |
| DC2 | 58505 | DC2 protein (Glycotransferase activity) | -2.12 | |
| GSTA1 | 2938 | Glutathione S-transferase A1 | -2.15 | |
| GAPD | 2597 | Glyceraldehyde-3-phosphate dehydrogenase | -2.16 | |
| GCLC | 2729 | Glutamate-cysteine ligase, catalytic subunit | -2.21 | |
| SRM | 6723 | Spermidine synthase | -2.39 | |
| HSP90AA1 | 3320 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | -2.46 | |
| AHCY | 191 | S-adenosylhomocysteine hydrolase | -2.48 | |
| MAT2A | 4144 | Methionine adenosyltransferase II, alpha | -2.74 | |
| Category 6: Cell physiology | NCF4 | 4689 | Neutrophil cytosolic factor 4, 40 kDa | 2.39 |
| CYCS | 54205 | Cytochrome c, somatic | -2.02 | |
| DBI | 1622 | GABA receptor modulator, acyl-Coenzyme A binding protein | -2.25 | |
| ATP5G3 | 518 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 | -2.26 | |
| HSPA1L | 3305 | Heat shock 70 kDa protein 1-like | -2.26 | |
| Category 7: Cell proliferation | FOSL1 | 8061 | FOS-like antigen 1 (transcription factor activity) | -2.09 |
| FGG | 2266 | Fibrinogen gamma chain | -2.36 | |
| FGG | 2244 | Fibrinogen beta chain | -2.45 | |
| Category 8: Cell adhesion | NELL2 | 4753 | NEL-like 2 (Calcium ion binding) | 2.25 |
| ITGB3 | 3690 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 2.11 | |
| RHOB | 388 | Ras homolog gene family, member B | -2.19 | |
| ADRM1 | 11047 | Adhesion regulating molecule 1 | -2.25 | |
| Category 9: Ubiquitination | UBE2N | 7334 | Ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | -2.02 |
| UBE2S | 27338 | Ubiquitin-conjugating enzyme E2S | -2.05 | |
| ANAPC7 | 51434 | Anaphase promoting complex subunit 7 | -2.15 | |
| CACYBP | 27101 | Calcyclin binding protein | -2.18 | |
| UBA52 | 7311 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | -2.29 | |
| COL6A3 | 1293 | Collagen, type VI, alpha 3 | 2.33 | |
| RPS3A | 6189 | Ribosomal protein S3A | -2.08 | |
| TWF1 | 5756 | Twinfilin, actin-binding protein, homolog 1 (Tyrosin kinase) | -2.01 | |
| AKR1C1 | 1645 | Aldo-keto reductase 1, member C1 (Bile acid binding) | -2.09 |
| Gene symbol | Gene ID (NCBI) | Gene name | Gene role | Primer | Primer sequence 5’-3’ |
| BMF | 90427 | Bcl2 modifying factor, transcript variant 1 | Has a single Bcl2 homology domain 3 (BH3), binds Bclk2 proteins and functions as an apoptotic activator | F | GCTTCAGTTGCATTGCAGACCAGTT |
| R | AGAGCCCTTGGGAATTCTCACCAT | ||||
| CD248 | 57124 | CD248 antigen = endosialin | A gene regulated by the cell density in vitro. Has a calcium binding domain | F | TCAACTACGTTGGTGGCTTCGAGT |
| R | AGTTGGGATAATGGGAAGCTGGGT | ||||
| PPM1E | 22843 | Protein phosphatase 1E | Member of the PP2C family of Ser/Thr phosphatases known to be negative regulators of stress response pathways | F | ATGCCTCCATTCACCTCCACGTTA |
| R | TGTCATAGAAGCCATCACAGGCCA | ||||
| FXYD3 | 5349 | FXY domain containing ion transport reg. 3 | The protein encoded by this gene may function as a chloride channel or as a chloride channel regulator | F | AATGCAAGTTTGGCCAGAAGTCCG |
| R | TTGCATATGAGGTCCCATGGCTGA | ||||
| OAS2 | 4939 | 2’-5’-oligoadenylate synthetase 2 | This enzyme family plays a significant role in the inhibition of cellular protein synthesis | F | AGAAGCCAACGTGACATCCTCGAT |
| R | TGCTGGAGTTCAGTGAAGCAGACT | ||||
| FY | 2532 | Duffy blood group antigen | Helps in leukocyte recruitment to sites of inflammation by facilitating movement of chemokines across the endothelium | F | TGACTCTGCACTGCCCTTCTTCAT |
| R | TTGACAACAGCAACAGCTTGGACC | ||||
| CERK | 64781 | Ceramide kinase | Integral to membranes, has roles in arachidonic acid release and production of eicosanoids | F | TGAGAAGAAACGGTGGTTGGGTCT |
| R | AGCATTTCCGGATGAGGATGAGGT | ||||
| HPSE | 10855 | Heparanase | Cell surface expression and secretion markedly promote tumor angiogenesis and metastasis | F | ACCTTTGCAGCTGGCTTTATGTGG |
| R | CTTGCACGCTTGCCATTAACACCT | ||||
| GAPDH | 2597 | Glyceraldehyde-3-phosphate dehydrogenase | Used as reference | F | GACCACAGTCCATGCCATCAC |
| R | GAGCTTCAGAAAGTGGTCGTTGA |
There were 21 genes influenced by two different treatment conditions (Table 1). Seventeen genes were affected by E. coli and L. plantarum, and four by E. coli and the combination of bacteria. Genes nos. 1-7 were up-regulated by both E. coli and L. plantarum; and genes nos. 8-17 were down-regulated by both bacteria. For each of the 17 genes in this group, the effects of the individual bacteria were brought to baseline by the combination treatment. In contrast, for three genes BMF, CD248 and PPM1E (nos. 18, 19 and 20 in Table 1), the stimulatory effect of E. coli was maintained after mixed infection with L. plantarum. For one gene (no. 21, CARD8), the 3.26-fold down-regulation by E. coli was reversed in the mixed infection, with demonstration of a four fold increase.
Apart from the specific up- and down-regulation of genes by either E. coli or L. plantarum, and reversal of E. coli-induced effects when L. plantarum was used as a co-infectant, several genes of physiologic importance were noted in our system. Table 2 describes 58 genes under 10 specific categories that were expressed during mixed infection. While the function of a small number of genes was not very well defined, most of the genes could be grouped into important cellular functions. These include genes involved in transcription regulation, RNA processing, protein biosynthesis, and other important processes such as ubiquitination, cell adhesion, proliferation and apoptosis.
Eight genes were randomly tested by quantitative real-time PCR to verify the expression detected by microarray (Table 3). For each of these genes, RT-PCR confirmed their expression after the three bacterial treatments in the same direction (stimulation or inhibition) as in the microarray experiments (Figure 2).
The infant gut is essentially sterile at birth and is first colonized with Enterobacteriaceae, which change the redox potential in the intestine and allow more microaerophilic and anaerobic species to colonize[42,43]. The latter group, which is comprised primarily of Bifidobacteria and Lactobacillus organisms[44], are considered as normal flora that coexist in the human colon, as new species are introduced to ultimately provide a stable flora in the human gut[45], in which over 800 bacterial species coexist in harmony[46]. In such a healthy state, the intestinal mucosa serves as the first line of defense against infections by providing an important mechanical and immunologic barrier between the host’s internal milieu and the gut environment. These intestinal epithelial cells generate and transmit signals between bacteria and deeper layers in the intestine[47]. In the event of specific infections, epithelial cells express and secrete proinflammatory and chemoattractant cytokines[48] that further transmit signals to the underlying cells in the reticuloendothelial system[47]. The virulence factors and the host responses to these factors in various diseases have been studied in a fair amount of detail (E. coli, Vibrio cholerae, Salmonella and Pseudomonas) using tissue culture and in vivo models, and specific genes and gene functions have been described[49-52]. These experiments have utilized single bacterial strains.
In an attempt to mimic the natural gut environment, communication systems among bacteria have also been studied relatively well. Chemical signals produced and detected by bacteria can be directed at other bacteria and self. This phenomenon, called as quorum sensing, is important for the microorganism’s adaptation to the local environment[53]. This fundamental prokaryotic behavior (among bacteria) is known to affect the symbiotic or antagonistic environment created within the gut milieu. However, the effect of single versus multiple bacterial species on eukaryotic cells has not been addressed in the literature.
The stimulus for us to conduct the current study came from our observation that a large number of probiotic trials have been conducted and reported in the recent past, with almost no basis for selection of the strain, and more importantly, with no data on changes in physiologic or pathologic parameters in the host, other than analysis of the primary and secondary clinical endpoints. Although a live bacterial supplement was used in all of these reported studies, there was also a serious lack of data on the colonizing ability of the probiotic strain and changes in the colonization by other bacteria in the host gut. Since the newborn gut is colonized with a paucity of bacteria (an average 2.5 species in preterm infants)[37,54] that expands to a limited but heterogenous flora by 10 d of age[55], we designed the current simple system to examine the effects of L. plantarum, a common human probiotic strain, and E. coli, the most common colonizing strain in the neonatal period, on gut cells. We took advantage of a microarray chip that allowed us to examine 19200 human genes in this simulated microbial gut environment. In this in vitro model, single and combined bacteria were allowed to interact with cultured cells, and our results were analyzed under high-stringency conditions to identify specific genes expressed during defined bacteria-gut cell interactions.
In our system, we observed a change (up- or down-regulation) in the expression of 333, 81 and 86 genes upon infection with E. coli, L. plantarum and the combined treatment, respectively. Our real-time PCR experiments confirmed the modifications demonstrated in the microarray experiments, albeit at a lower level, a phenomenon also reported in other studies[50]. The numbers of unique genes presented in this study are in the range reported in previous studies in which Gram-negative enteric pathogens modified the expression of 0.5%-13% of the genes in epithelial cells[50,56-58], and commensal bacteria induced differential expression of 0.35%-6.2% of examined genes in mouse colonocytes[59]. Our strain of E. coli modified 1.73%, and L. plantarum modified 0.43% of genes. The slightly lower number of genes identified in our 19200 array may have been due to the use of a non-pathogenic strain of E. coli, a commensal Lactobacillus, and an array that included only constitutively expressed genes. Genes expected to be expressed after a bacterial insult such as pro-inflammatory cytokines were not spotted on this array. Additionally, a slightly low number might have resulted from our conservative choice of a twofold increase in expression as being significant in our analysis.
There are several comparisons that can be made between our results and those of others using a similar approach but with single bacterial infection. For example, from the six genes up-regulated by enteropathogenic E. coli in HeLa cells[49], we found only one (zyxin, a cytoskeletal protein) to be in common with our microarray results. There was a similar increase (1.72-fold) in expression of this gene when our E. coli strain 6-1 was used to infect Caco-2 cells. Two previous studies with commensal flora have reported that bacterial reconstitution of germ-free mice increased the expression of the colon-specific serum amyloid A1 gene[60,61]. In our model, serum amyloid A2 gene expression was increased by 2.22-fold. From the 12 genes down-regulated by non-pathogenic bacterial reconstitution of germ-free mice, reported by Fukushima et al[59] in colonic epithelial cells, three were in common with our microarray; selenoprotein P, 3-hydroxy-3 methylglutaryl-coenzyme A synthase and metallothionein. All three were also down-regulated in our combination treatment model. The authors also showed a down-regulation of solute carrier family 20 - member 1. Our results were very similar to this observation in that we also noted a decrease in the expression of other members of the solute carrier families, i.e., family 2, 9, 12, 20, 24, 25 and 35. Fukushima et al[59] have shown overexpression of heat shock protein (60 kDa) in germ-free mice compared to specific pathogen-free rodents that had received treatment with normal mouse flora. We observed a similar phenomenon in our system in which down-regulation of heat shock proteins 75, 105 and an ortholog of mouse heat shock protein 70 kDa were noted after combined bacterial treatment. We observed cytochrome c oxidase subunits IV isoform 1, Va, VIb, VIc, VIIa, VIIb, VIIc and VIII to be up-regulated after L. plantarum treatment, similar to that described by Hooper et al, who demonstrated up-regulation of cytochrome c oxidase subunit 1 by Bacteroides, another species also considered as commensal flora. Hooper and colleagues have also shown up-regulation of calmodulin after treatment with Bacteroides[61]. Similar increases in expression were noted for calmodulin 1, 2 and 3, calmodulin-dependent protein kinase and phosphodiesterase in our system.
We observed modulation of multiple genes known to have an impact on cellular and physiologic processes in the eukaryotic system (Table 2). These genes ranged from basic transcriptional regulators to those involved in protein synthesis, cellular metabolism, cell proliferation and apoptosis. During mixed infection, we observed down-regulation of three genes involved in ubiquitination. Ubiquitin-conjugating enzyme E2N, ubiquitin-carrier protein E2-EPF and ubiquitin A-52 residue ribosomal protein fusion product 1 were reduced 2.02, 2.05 and 2.29-fold, respectively. In a recent study that investigated anti-inflammatory properties of Lactobacillus casei, expression of several genes involved in ubiquitination was reduced, including E2N, a gene (common to our system) that was reported to be decreased 2.88-fold[62]. The authors concluded I-κB stabilization via reduced ubiquitination and downstream modulation of inflammatory response driven by NF-κB in Shigella-infected Caco-2 cells. We used a non-pathogenic commensal strain of E. coli in our experiments, and while the aim of the current study was not to assess or examine the effects of L. plantarum during bacterial infection or inflammation, our results strongly suggest that Lactobacillus strains do indeed affect common physiologic pathways in gut cells, which may ultimately shape the host response in health and disease.
In our study, it was important and intriguing to note that the three experimental infections induced quite unique gene-expression profiles. Even the mixed infection with E. coli and L. plantarum had a very small overlap with the expression profiles of the strains when they were used alone. This illustrates how colonization can change the gene expression of host cells as they are exposed to more than one species of bacteria. In real life, the gut cells are exposed to a multitude of bacterial strains, and hence, it may be of limited value to study the effect of infection or colonization by single bacterial species in a clean tissue culture environment, and use the results as the basis for designing treatment or preventive strategies. Using neonatal models of gut colonization, we have previously shown that bacterial ecology (combination of Gram-negative and Gram-positive organisms), rather than individual virulent bacterial strains, plays a more important role in diseases such as NEC[36]. The results of our current study are in line with previous observations, and now provide an additional line of support and offer a possible explanation for the varied results of recent probiotic trials. On a broader scale, this report provides an insight into the complex host response that can be expected at mucosal sites such as the gastrointestinal tract. Based on the results obtained from tissue culture with only two bacteria in the system, it can be speculated that our findings are only the tip of the iceberg, and the real in vivo picture in mammals will be even more complex. While it is becoming increasing clear that specific Lactobacillus species posses unique health-promoting characteristics[29], knowledge gained from the current study further indicates that a "one strain fits all" approach may not always succeed in the treatment or prevention of specific diseases. A more global approach needs to be taken with proper emphasis on the microbial ecology, while addressing the pathogenesis of unique bacterial diseases in the mammalian intestine at different ages and stages of development.
In the context of in vivo or clinical trial environment, it should be noted that our current model and results do not represent a universal phenomenon, nor provide a comprehensive picture of the human intestine. For example, genes expressed will probably be different if other probiotic strains such as Bifidobacteria and L. casei were used in our system. Similarly, combinations of other aerobic and anaerobic Gram-negative and Gram-positive strains may induce different sets of genes. We can utilize other microarray systems with cytokine and signaling-molecule genes (not spotted in the current 19200 gene array), when our aim would be to identify modifications in inflammatory mediators. The relative concentrations of each bacterium in the system may also change the gene-expression profile. In the current study, we selected a 1:10 ratio of E. coli to Lactobacillus infecting dose to simulate the human intestinal microflora, in which anaerobic and microaerophilic organisms form the dominant flora[63]. Since enteric bacteria such as E. coli are sometimes present at < 0.1% of the total bacterial population, with a predominance by obligate anaerobes[64], it is not unexpected to observe a different gene-expression profile when a 10-100-fold higher proportion of Lactobacilli are used in the system. Nevertheless, such manipulations and experiments can be done, and despite some limitations, assessment of mRNA-expression profiles by cDNA array analysis can be utilized as a useful technique for expanding our understanding of the colonocyte-bacteria interaction[50].
While it may appear difficult to analyze complex microflora (400-800 species) and their interactions with gut cells in the mature intestine, this is now made feasible with the availability of new techniques. Fluorescent in situ hybridization utilizing bacterial rRNA can identify and quantify major genera of bacteria, even if they are non-culturable in stools[65,66]. Bacterial microarray chips developed during the last year can identify thousands of bacterial species in stools in one experiment[67,68]. Denaturing gradient gel electrophoresis can be utilized to monitor changes in microflora pattern[69,70] over time and after administration of probiotic supplements. Live colonocytes can be isolated from stool samples and used to examine the expression of genes and proteins during different experimental and/or disease states[71,72]. At this juncture, there is a need for the scientific community to engage in careful evaluation of probiotic strains in in vitro and in vivo systems prior to initiation of clinical trials. With the new non-invasive tools at hand, such preclinical endeavors, coupled with concurrent examination of changes in the gut flora and host responses during clinical trials, hold great promise in discerning the difference between "snake oil" and "magic bullets" when it comes to the role of probiotic therapy in human medicine.
S- Editor Liu Y L- Editor Kerr C E- Editor Liu Y
| 1. | Atrih A, Rekhif N, Michel M, Lefebvre G. Detection of bacteriocins produced by Lactobacillus plantarum strains isolated from different foods. Microbios. 1993;75:117-123. [PubMed] |
| 2. | Bernet MF, Brassart D, Neeser JR, Servin AL. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35:483-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 473] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 3. | Perdigón G, Maldonado Galdeano C, Valdez JC, Medici M. Interaction of lactic acid bacteria with the gut immune system. Eur J Clin Nutr. 2002;56 Suppl 4:S21-S26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 224] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 4. | Gordon JI, Hooper LV, McNevin MS, Wong M, Bry L. Epithelial cell growth and differentiation. III. Promoting diversity in the intestine: conversations between the microflora, epithelium, and diffuse GALT. Am J Physiol. 1997;273:G565-G570. [PubMed] |
| 5. | Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1598] [Cited by in RCA: 1560] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 6. | Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1139] [Cited by in RCA: 1078] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 7. | Reid G, Bruce AW, McGroarty JA, Cheng KJ, Costerton JW. Is there a role for lactobacilli in prevention of urogenital and intestinal infections? Clin Microbiol Rev. 1990;3:335-344. [PubMed] |
| 8. | de Waard R, Garssen J, Bokken GC, Vos JG. Antagonistic activity of Lactobacillus casei strain shirota against gastrointestinal Listeria monocytogenes infection in rats. Int J Food Microbiol. 2002;73:93-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 9. | Taylor CJ, Mahenthiralingam E. Functional foods and paediatric gastro-intestinal health and disease. Ann Trop Paediatr. 2006;26:79-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Szajewska H, Setty M, Mrukowicz J, Guandalini S. Probiotics in gastrointestinal diseases in children: hard and not-so-hard evidence of efficacy. J Pediatr Gastroenterol Nutr. 2006;42:454-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 96] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Sazawal S, Hiremath G, Dhingra U, Malik P, Deb S, Black RE. Efficacy of probiotics in prevention of acute diarrhoea: a meta-analysis of masked, randomised, placebo-controlled trials. Lancet Infect Dis. 2006;6:374-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 294] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 12. | Saiman L. Strategies for prevention of nosocomial sepsis in the neonatal intensive care unit. Curr Opin Pediatr. 2006;18:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Novak J, Katz JA. Probiotics and prebiotics for gastrointestinal infections. Curr Infect Dis Rep. 2006;8:103-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Johnston BC, Supina AL, Vohra S. Probiotics for pediatric antibiotic-associated diarrhea: a meta-analysis of randomized placebo-controlled trials. CMAJ. 2006;175:377-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Duchmann R. The role of probiotics and antibiotics in regulating mucosal inflammation. Adv Exp Med Biol. 2006;579:219-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Doron S, Gorbach SL. Probiotics: their role in the treatment and prevention of disease. Expert Rev Anti Infect Ther. 2006;4:261-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Daniel C, Repa A, Wild C, Pollak A, Pot B, Breiteneder H, Wiedermann U, Mercenier A. Modulation of allergic immune responses by mucosal application of recombinant lactic acid bacteria producing the major birch pollen allergen Bet v 1. Allergy. 2006;61:812-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Chan-Yeung M, Becker A. Primary prevention of childhood asthma and allergic disorders. Curr Opin Allergy Clin Immunol. 2006;6:146-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Boyle RJ, Tang ML. The role of probiotics in the management of allergic disease. Clin Exp Allergy. 2006;36:568-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Kalliomäki MA, Isolauri E. Probiotics and down-regulation of the allergic response. Immunol Allergy Clin North Am. 2004;24:739-752, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Miraglia del Giudice M, De Luca MG. The role of probiotics in the clinical management of food allergy and atopic dermatitis. J Clin Gastroenterol. 2004;38:S84-S85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Kitajima H, Sumida Y, Tanaka R, Yuki N, Takayama H, Fujimura M. Early administration of Bifidobacterium breve to preterm infants: randomised controlled trial. Arch Dis Child Fetal Neonatal Ed. 1997;76:F101-F107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 231] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 23. | Hoyos AB. Reduced incidence of necrotizing enterocolitis associated with enteral administration of Lactobacillus acidophilus and Bifidobacterium infantis to neonates in an intensive care unit. Int J Infect Dis. 1999;3:197-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 234] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Lin HC, Su BH, Chen AC, Lin TW, Tsai CH, Yeh TF, Oh W. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2005;115:1-4. [PubMed] |
| 25. | Bin-Nun A, Bromiker R, Wilschanski M, Kaplan M, Rudensky B, Caplan M, Hammerman C. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. 2005;147:192-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 364] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 26. | Dani C, Biadaioli R, Bertini G, Martelli E, Rubaltelli FF. Probiotics feeding in prevention of urinary tract infection, bacterial sepsis and necrotizing enterocolitis in preterm infants. A prospective double-blind study. Biol Neonate. 2002;82:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 267] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 27. | Agarwal R, Sharma N, Chaudhry R, Deorari A, Paul VK, Gewolb IH, Panigrahi P. Effects of oral Lactobacillus GG on enteric microflora in low-birth-weight neonates. J Pediatr Gastroenterol Nutr. 2003;36:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 28. | Madsen KL. The use of probiotics in gastrointestinal disease. Can J Gastroenterol. 2001;15:817-822. [PubMed] |
| 29. | Isolauri E. Probiotics in human disease. Am J Clin Nutr. 2001;73:1142S-1146S. [PubMed] |
| 30. | Vanderhoof JA, Young RJ. Probiotics in pediatrics. Pediatrics. 2002;109:956-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Reece RJ, Beynon L, Holden S, Hughes AD, Rébora K, Sellick CA. Nutrient-regulated gene expression in eukaryotes. Biochem Soc Symp. 2006;85-96. [PubMed] |
| 32. | Cashin P, Goldsack L, Hall D, O'Toole R. Contrasting signal transduction mechanisms in bacterial and eukaryotic gene transcription. FEMS Microbiol Lett. 2006;261:155-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 33. | Raupach B, Mecsas J, Heczko U, Falkow S, Finlay BB. Bacterial epithelial cell cross talk. Curr Top Microbiol Immunol. 1999;236:137-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 34. | Ahrné S, Nobaek S, Jeppsson B, Adlerberth I, Wold AE, Molin G. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol. 1998;85:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 309] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 35. | Engle MJ, Goetz GS, Alpers DH. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J Cell Physiol. 1998;174:362-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 36. | Panigrahi P, Gupta S, Gewolb IH, Morris JG. Occurrence of necrotizing enterocolitis may be dependent on patterns of bacterial adherence and intestinal colonization: studies in Caco-2 tissue culture and weanling rabbit models. Pediatr Res. 1994;36:115-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 39] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Gupta S, Morris JG, Panigrahi P, Nataro JP, Glass RI, Gewolb IH. Endemic necrotizing enterocolitis: lack of association with a specific infectious agent. Pediatr Infect Dis J. 1994;13:728-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 38. | Panigrahi P, Bamford P, Horvath K, Morris JG, Gewolb IH. Escherichia coli transcytosis in a Caco-2 cell model: implications in neonatal necrotizing enterocolitis. Pediatr Res. 1996;40:415-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Ichikawa JK, Norris A, Bangera MG, Geiss GK, van 't Wout AB, Bumgarner RE, Lory S. Interaction of pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc Natl Acad Sci USA. 2000;97:9659-9664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 135] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Ichikawa JK; DeRisi. Amino-alyl Dye Coupling Protocol. Available from: http: //derisilab.ucsf.edu/pdfs/amino-allyl-protocol.pdf. 2001. |
| 41. | Hasseman J. Microarray labeled probe hybridization. Available from: http: //www.tigr.org/tdb/microarray/protocolsTIGR.shtml. 2002. |
| 42. | Mai V, Morris JG. Colonic bacterial flora: changing understandings in the molecular age. J Nutr. 2004;134:459-464. [PubMed] |
| 43. | Freter R. Control mechanisms of the large-intestinal microflora and its influence on the host. Acta Gastroenterol Latinoam. 1989;19:197-217. [PubMed] |
| 44. | Orrhage K, Nord CE. Bifidobacteria and lactobacilli in human health. Drugs Exp Clin Res. 2000;26:95-111. [PubMed] |
| 45. | Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 1468] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 46. | Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3394] [Cited by in RCA: 3545] [Article Influence: 177.3] [Reference Citation Analysis (5)] |
| 47. | Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 436] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 48. | Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 881] [Cited by in RCA: 881] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 49. | de Grado M, Rosenberger CM, Gauthier A, Vallance BA, Finlay BB. Enteropathogenic Escherichia coli infection induces expression of the early growth response factor by activating mitogen-activated protein kinase cascades in epithelial cells. Infect Immun. 2001;69:6217-6224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J Biol Chem. 2000;275:14084-14094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Firoved AM, Deretic V. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol. 2003;185:1071-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 113] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 52. | Stokes NR, Zhou X, Meltzer SJ, Kaper JB. Transcriptional responses of intestinal epithelial cells to infection with Vibrio cholerae. Infect Immun. 2004;72:4240-4248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 53. | Konaklieva MI, Plotkin BJ. Chemical communication--do we have a quorum? Mini Rev Med Chem. 2006;6:817-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 54. | Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80:F167-F173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 251] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 55. | Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317-321. [PubMed] |
| 56. | Belcher CE, Drenkow J, Kehoe B, Gingeras TR, McNamara N, Lemjabbar H, Basbaum C, Relman DA. The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter-defensive strategies. Proc Natl Acad Sci USA. 2000;97:13847-13852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 57. | Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J Immunol. 2000;164:5894-5904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 157] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 58. | Pédron T, Thibault C, Sansonetti PJ. The invasive phenotype of Shigella flexneri directs a distinct gene expression pattern in the human intestinal epithelial cell line Caco-2. J Biol Chem. 2003;278:33878-33886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 59. | Fukushima K, Ogawa H, Takahashi K, Naito H, Funayama Y, Kitayama T, Yonezawa H, Sasaki I. Non-pathogenic bacteria modulate colonic epithelial gene expression in germ-free mice. Scand J Gastroenterol. 2003;38:626-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 60. | Ogawa H, Fukushima K, Sasaki I, Matsuno S. Identification of genes involved in mucosal defense and inflammation associated with normal enteric bacteria. Am J Physiol Gastrointest Liver Physiol. 2000;279:G492-G499. [PubMed] |
| 61. | Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1565] [Cited by in RCA: 1495] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 62. | Tien MT, Girardin SE, Regnault B, Le Bourhis L, Dillies MA, Coppée JY, Bourdet-Sicard R, Sansonetti PJ, Pédron T. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228-1237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 63. | Onderdonk AB. The intestinal microflora and intra-abdominal sepsis. Medical importance of the normal microflora. Dordrecht, Boston, London: Kluwer Academic Publishers 1999; 164-176. [DOI] [Full Text] |
| 64. | Evaldson G, Heimdahl A, Kager L, Nord CE. The normal human anaerobic microflora. Scand J Infect Dis Suppl. 1982;35:9-15. [PubMed] |
| 65. | Harmsen HJ, Raangs GC, He T, Degener JE, Welling GW. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68:2982-2990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 271] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 66. | Mai V, Katki HA, Harmsen H, Gallaher D, Schatzkin A, Baer DJ, Clevidence B. Effects of a controlled diet and black tea drinking on the fecal microflora composition and the fecal bile acid profile of human volunteers in a double-blinded randomized feeding study. J Nutr. 2004;134:473-478. [PubMed] |
| 67. | Kostić T, Weilharter A, Rubino S, Delogu G, Uzzau S, Rudi K, Sessitsch A, Bodrossy L. A microbial diagnostic microarray technique for the sensitive detection and identification of pathogenic bacteria in a background of nonpathogens. Anal Biochem. 2007;360:244-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Palmer C, Bik EM, Eisen MB, Eckburg PB, Sana TR, Wolber PK, Relman DA, Brown PO. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 2006;34:e5. [PubMed] |
| 69. | Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 246] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 70. | Suau A. Molecular tools to investigate intestinal bacterial communities. J Pediatr Gastroenterol Nutr. 2003;37:222-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 71. | Nair P, Lagerholm S, Dutta S, Shami S, Davis K, Ma S, Malayeri M. Coprocytobiology: on the nature of cellular elements from stools in the pathophysiology of colonic disease. J Clin Gastroenterol. 2003;36:S84-S93; discussion S94-S96. [PubMed] |
| 72. | Matsushita H, Matsumura Y, Moriya Y, Akasu T, Fujita S, Yamamoto S, Onouchi S, Saito N, Sugito M, Ito M. A new method for isolating colonocytes from naturally evacuated feces and its clinical application to colorectal cancer diagnosis. Gastroenterology. 2005;129:1918-1927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |