Published online Mar 7, 2006. doi: 10.3748/wjg.v12.i9.1452
Revised: October 1, 2005
Accepted: October 26, 2005
Published online: March 7, 2006
AIM: To develop a highly efficacious method for preparation of soluble SARS S-protein using adenovirus vector to meet the requirement for S-protein investigation.
METHODS: The human adenovirus vector was used to express the soluble S-protein (corresponding to 1~1190 amino acids) fused with Myc/His tag using codon-optimized gene construct in HEK239 cells. The recombinant adenovirus bearing S-protein gene was generated by ligation method. The expressed S-protein with Myc/His tag was purified from culture medium with Ni-NTA agarose beads followed by dialysis. The S-protein was detected by Western blot and its biologic activity was analyzed by binding to Vero cells.
RESULTS: Under the conditions of infection dose (MOI of 50) and expression time (48 h), the high-level expression of S-protein was obtained. The expression level was determined to be approximately 75 μg/106 cells after purification. Purified soluble S-protein was readily detected by Western blot with anti-Myc antibody and showed the ability to bind to surface of Vero cells, demonstrating that the soluble S-protein could remain the biologic activity in the native molecule.
CONCLUSION: The high-level expression of S-protein in HEK293 cells mediated by adenovirus can be achieved under the optimized expression conditions. The proteins possess the biologic activity, which lays a foundation for further investigation of S-protein biological function.
- Citation: Zhong F, Zhong ZY, Liang S, Li XJ. High expression level of soluble SARS spike protein mediated by adenovirus in HEK293 cells. World J Gastroenterol 2006; 12(9): 1452-1457
- URL: https://www.wjgnet.com/1007-9327/full/v12/i9/1452.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i9.1452
Severe acute respiratory syndrome (SARS) is a life-threatening disease. The etiological agent of SARS has been identified as a novel coronavirus, called SARS coronavirus (SARS-CoV)[1-3]. SARS-CoV is a positive-strand RNA virus encoding several structural proteins including spike (S) glycoprotein, nucleocapsid protein (N), membrane protein (M) and envelope protein (E)[4,5]. S-protein, a 1255 amino-acid type I membrane glycoprotein, is the prominent protein present in viral membrane and consists of a signal peptide sequence (amino acids 1 to 13), an extracellular domain (amino acid 14 to 1190), a transmembrane domain (amino acids 1191 to 1227) and a short intracellular tail (amino acids 1228 to 1255). S-protein plays an important role in SARS coronavirus infection by interaction with cellular receptor, angiotensin-converting enzyme 2 (ACE2) on the host cells[6-8]. S-protein also has efficient antigenicity and could induce neutralization antibodies[9-12]. These characteristics make it a potential candidate for subunit vaccine and anti-SARS drug development and diagnostic application. Large-scale preparation of S protein is desirable for S protein investigation, such as protein-protein interaction, S protein-based vaccination and diagnostic approach. In order to obtain sufficient quantities of S-protein, we used human adenovirus vector to construct the recombinant adenovirus bearing SARS soluble S-protein gene and express it in human embryonic kidney 293 cells (HEK293 cells).
HEK293, HEK293T and Vero African green monkey kidney cells were purchased from American Type Culture Collection (ATCC); Manassas VA. HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 100 u/mL penicillin, 100 μg/mL streptomycin, 2mM L-glutamine and 1×MEM non-essential amino acid solution ( DMEM complete medium) at 37°C and 50 mL/L CO2. Vero cells were cultured in medium 199 supplemented with the same components as above except for 1×MEM non-essential amino acid solution (medium-199 complete medium). Adenovirus vectors (pShuttle2 and Adeno-X DNA) based on human adenovirus type 5 (Ad5) were purchased from BD Sciences Clontech (Palo Alto, CA). QuikChange® XL site-directed mutagenesis kit was obtained from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were obtained from Promega (Madison, WI).
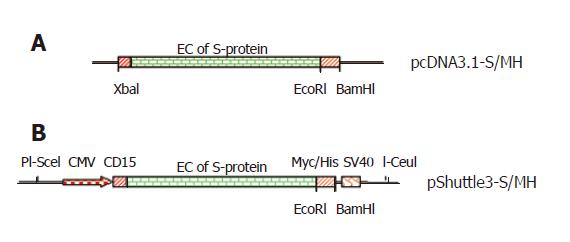
The full-length codon-optimized SARS-CoV S-protein cDNA, cloned into XbaI/BamHI sites in the pcDNA3.1 (-) expression vector, was kindly provided by Dr. Mecheal Farzan (Brigham and Women’s Hospital, Harvard University, Boston, USA) [6]. The vector was modified to express soluble SARS S-protein fused with Myc/His tag at C-terminus. Briefly, a EcoRI site was introduced in S-protein DNA between extracellular and transmembrane domains (corresponding to residues K1193 and W1194) using QuikChange® XL site-directed mutagenesis kit and the following primers: 5’gtacgagcagtacatcgaattcccttggtatgtgtggctg3’ and 5’ cagccacacataccaagggaattcgatgtaatgctcgtac3’. The EcoRI-mutated expression vector was then digested with EcoRI and BamHI. The DNA fragment encoding Myc/His with additional EcoRI and BamHI sticky ends at 5’ and 3’ terminuses was generated by annealing the following primers: 5’aattcgaacaaaaactcatctcagaagaggatctg(myc) accggtcatcatcaccatcaccat(6His)tga(stopcodon)g3’/5’gatcctca(stopcodon)atggtgacggtgatgatg(6His)accggtcagatcctc ttctgagatgagtttttgttc(myc)g3’. Then the Myc/His fragment was inserted into the EcoRI and BamHI sites of EcoRI-mutated vector to generate pcDNA3.1-S/MH containing the soluble S-protein fused with Myc/His gene.
In order to conveniently subclone fused soluble S-protein gene into adenoviral shuttle vector, the multiple cloning sites (MCS) of pShuttle2 were modified with a new MCS sequence. Briefly, the new MCS sequence was amplified by PCR method with a pair of primers: 5’tttggctagcggtaccgcggccgcggatccaagatatcgggc3’ and 5’tttcccgggttcgaatctagagggcccgatatcttggatc3’ under the conditions of 94 °C for 5 min, 50 °C for 1 min and 72 °C for 10 min. The amplified 65 bp PCR product (the new MCS sequence) was digested with SmaI (blunt end ) and NheI, respectively. The digested products were purified with GFXTM PCR DNA and gel band purification kit (Amersham). The pShuttle2 plasmid was first digested with KpnI, after filled using Klenow DNA polymerase to generate blunt end, the linearized plasmid was then digested with NheI. Finally, the new MCS was inserted into NheI and blunt sites of the pShuttle2 to generate pShuttle3 vector.
The fused soluble S-protein gene in pcDNA3.1-S/MH was cut out with XbaI and BamHI and downstream cytomegalovirus immediate early (CMV) promoter of pShuttle3 was subcloned using NheI and BamHI sites to produce pShuttle3-S/MH.
Recombinant adenovirus vector expressing fused soluble S-protein gene downstream of CMV promoter was constructed using the Adeno-XTM expression system (BD Sciences Clontech), following the manufacturer’s instructions by the ligation method (Mizuguchi & Kay 1998)[13]. Briefly, the pShuttle3-S/MH was digested with PI-SceI and I-CeuI to release the expression cassette including the promoter, fused soluble S-protein gene and the SV40 polyadenylation signal (SV40 poly A). This DNA fragment was subsequently ligated to the pAdeno-X DNA (provided with kit) to generate pAdeno-S/MH. After ligation it was digested with SwaI restriction enzyme to eliminate non-recombinant plasmids and used to transform E. coli DH5α competent cells, clones bearing ampicillin resistance were selected. Positive recombinant plasmids were identified by screening plasmids with XhoI digestion.
Approximately 1-2×106 HEK293 cells were plated in T25 flasks 24h before transfection when they reached 60-80% confluence. About 3 μg of pAdeno-S/MH, digested with PacI, was transfected into HEK293 cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. The transfected HEK293 cells were cultured in DMEM complete medium at 37 °C and 50mL/L CO2. After two days the medium was changed to maintaining medium (containing 5% serum). After about two-week maintaining culture, the cytopathic effect (CPE) appeared in the monolayer of HEK293 cells. The cells was harvasted and viral DNA was prepared with Hirt method[14]. The recombinant adenovirus carrying S-protein gene (Ad-S/MH) was identified with PCR method using a pair of primers: 5’agcaacttccgcgtg3’/5’ggtgttgcggtcctg3’ (corresponding to Ser301 to Thr760 residues) and the viral DNA as template. The viral titer was determined by plague assay in HEK 293 cells.
Eighty percent confluent HEK 293 cells in 4×T75 flasks were infected with 3 mL harvested medium from the initial recombinant adenovirus lysates (~10 MOI) in 20 mL culture medium. One day later, the medium was replaced with serum-free medium (Opti-MEM, 50μg/mL BSA, L-glutamine and antibiotics ). After 48h, the medium containing S-protein was harvested and centrifuged at 8 000 r/min for 10 min. The supernatant was mixed with Ni-NTA-agrose beads and 1×binding buffer (20 mmol/L Tris, 150 mmol/L NaCl, 5 mmol/L imidazole, pH 8.0), incubated at 4 °C overnight with shaking, and then precipitated. The beads were washed with washing buffer (20 mmol/L Tris, 500 mmol/L NaCl, 10 mmol/L imidazole, pH 8.0). Bound S-protein was eluted with elutting buffer (20 mmol/L Tris, 500 mmol/L NaCl, 250 mmol/L imidazole, pH 8.0).
The eluted samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5% (w/v) gels. The separated proteins were either stained with silver staining method or electro-blotted and detected by Western-blotting using a mouse anti-His antibody followed by a goat anti-mouse IgG conjugated with alkaline phosphatase. The blots were developed using the Immun-StarTM substrate pack (Bio-Rad).
The eluted protein was dialysed against PBS (pH 8.0) to elimminate imidazole. The protein concentrations were determined using the Bio-Rad (CA, USA) protein assay kit developed based on the method of Bradford (Bradford, 1976) [15].
The Vero and HEK293T cells were detached from flask walls with 0.5mM EDTA in PBS, washed and re-suspended in PBS containing 0.5% (w/v) BSA (PBS-BSA) at ~4×107 cells/mL. Each cell sample (50 μL ) was incubated with 2 μg of S-protein for 3 h at 4°C with rotation. The cells were washed with PBS-BSA and incubated in 20% (v/v) goat serum in PBS-BSA for 30 min on ice. After washing with cold PBS-BSA, the cells were incubated with a mouse anti-myc antibody (Roche, Indianapolis, OIN) at 10 μg/mL for 30 min on ice. As a control, the cells were not incubated with S-protein. After washing with PBS-BSA, the cells were incubated with goat anti-mouse IgG conjugated with PRE for 40 min on ice, were washed and fixed in 1% (w/v) paraformaldehyde and analyzed by a FACScalibur flow cytometer (BD Biosciences).
SARS-CoV S-protein, like other coronaviruses, is the prominent surface protein existed in viral lipid envelope, which mediates virus interaction with host cells leading to infection. To investigate the S-protein-mediated interaction between virus and host, soluble S-protein was needed for the investigations.
In order to express the codon-optimized soluble S-protein fused with Myc/His tag using adenovirus vector, the transmembrane and C-terminal domain of full-length S-protein gene were deleted by creating EcoRI site immediately downstream the extracellular domain and then the DNA fragment encoding Myc/His with stop codon was inserted into EcoRI and BamHI sites of the EcoRI-mutated vector to generate pcDNA3.1-S/MH ( Figure 1A). The pShuttle3-S/MH (Figure 1B) was constructed by transferring soluble S-protein gene fused with Myc/His tag from pcDNA3.1-S/MH into adenoviral pShuttle3 vector derived from pShuttle2 by modification on MCS. The fusion of Myc/His tag to S-protein could facilitate purification and detection of expressed S-protein.
The entire expression cassette of S-protein, which in pShuttle3-S/MH (Figure 1B) is flanked by unique PI-SceI and I-CeuI sites, was released using these enzymes and ligated into the pAdeno-X DNA vector. The new vector contained a different resistance marker (ampicillin) from pShuttle3-S/MH (kanamycin), preventing pShuttle3-S/MH recovery. The candidate clones were screened by digesting the recombinant plasmids using XhoI enzyme. Three positive pAdeno--S/MH plasmids were constructed.
The plasmid pAdeno-S/MH containing two PacI restriction sites, was located in both ends of the viral genome, 3’ and 5’ of the inverted terminal repeats (ITRs). Thus, to ensure efficient replication and packaging of recombinant adenovirus carrying S-protein gene (Ad-S/MH), the plasmid was digested with PacI and used to transfect HEK293 cells. After 15 d plaques appeard and then a large number of cells were detached from the flask, indicating the presence of adenovirus. To confirm the presence of the S-protein gene in recombinant adenovirus, the viral DNA was extracted and identified by PCR using the viral DNA as template. The specific DNA fragment (1 370 bp) was amplified from recombinant adenoviral DNA .
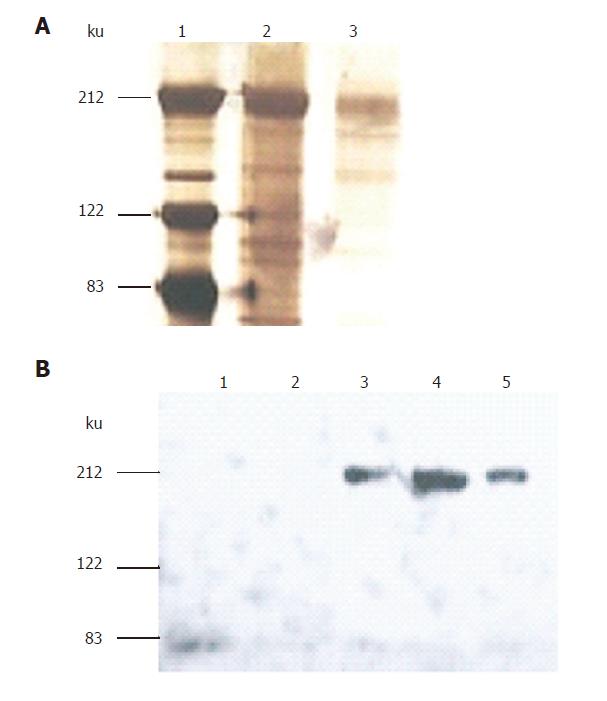
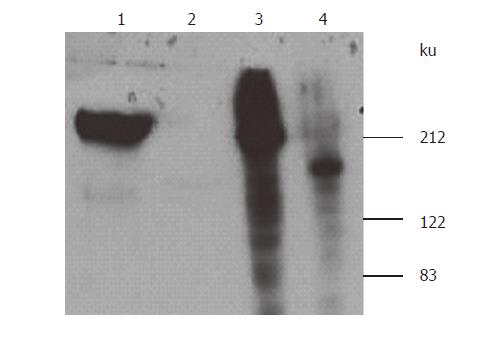
Using Ad-S/MH adenoviruses to infect HEK 293 cells cultured in serum-free medium, the medium was harvested. The S-protein was purified using Ni-NTA agarose columns binding to the C-terminal His tag at 4 °C. Fractions of eluted proteins were analyzed by SDS-PAGE under reducing conditions and identified either by silver staining and assignment using a molecular marker (Figure 2A) or Western-blotting using anti-Myc antibody detection (Figure 2B). The analysis revealed that the size of the purified S-protein under reducing conditions was consistent with previous studies[6,16]. However, it was much larger than the calculated molecular weight of ~132ku (1199 residues: 1181 residues of extracellular domain and 18 residues of myc/His tag (EQKLISEEDLTGHHHHHH)). This difference was considered to be related to glycosylation[16] since the extracellular domain of S-protein was predicted to have 18 potential N-link glycosylation sites, contributing to the observed such bigger molecular weight of S-protein.
Tunicamycin, an inhibitor of N-linked glycosylation, was used to treat the cells (7μg/mL) when they were infected with Ad-S/MH adenoviruses. S-protein secretion from the cells into the media was effectively blocked in the presence of tunicamycin, S-protein could not be detected by Western blotting from the culture media. In contract, without tunicamycin, S-protein could be detected from the media. However, when the cell lysate was detected by Western blotting, S-protein was detected from tunicamycin-treated cells as a size of 130 ku, smaller than the size of ~200 ku detected from non-tunicamycin-treated cells (Figure 3). This indicated that the low molecular weight of S-protein in tunicamycin-treated cells was due to lack of N-liked glycosylation, interfering with S-protein maturation and secretion.
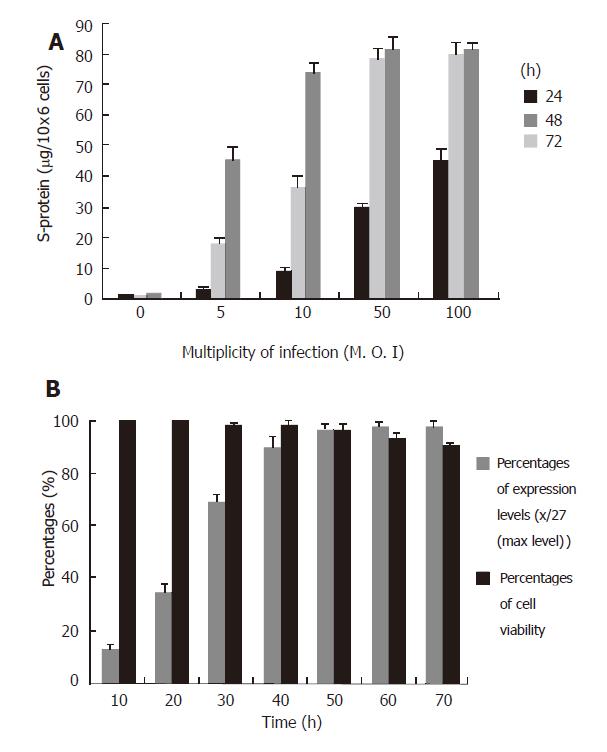
To determine the optimal dose (M.O.I.) of adenovirus for cell infection and optimal time for harvesting the culture media for S-protein purification, the HEK293 cells were prepared at the same confluence (about 70%) and infected with different doses of Ad-S/MH viruses and the culture media were harvested at different time points. The purified S-protein levels were evaluated with Bio-Rad protein assay kit. The high-level S-protein (above 75μg/106 cells) was obtained using an MOI of 10-100 at 72 h, or 50-100 at 48 h after infection (Figure 4A). On the basis of these results, we chose an MOI of 50 for viral infection and 48 h for expression time as the suitable conditions because these conditions led to high-level expression and low time-consuming. To confirm these conditions, time-course of expression and cell viabilities were carried out with MOI of 50 for infection. The high expression levels (above 75 µg/106 cells) and high cell viabilities (above 96%) were obtained using MOI of 50 between 40 h and 50 h periods (Figure 4B). We repeated three expressions and purifications under these conditions. The average yield of purified S-protein was approximately 75 μg/106 cells.
ACE2 of host cells was reported to be a functional receptor for SARS-CoV[6]. S-protein interaction with ACE2 was reported to be related to S-protein extracellular domain, in which amino acids 270 to 510 were identified to be essential for the interaction[17,18]. Vero cells, but not 293T cells could permit replication of SARS-CoV since the receptor ACE2 could be expressed on Vero but not 293T cells[6]. To determine whether the expressed S-protein had the biological activity to bind to Vero cells, Vero cells and 293T cells (as a negative control) were incubated with the purified S-protein and bound S-protein was detected by flow cytometry using a mouse anti-Myc antibody. Binding assay showed that the purified S-protein could bind to Vero cells (Figure 5A) but not 293T cells (Figure 5B). These results demonstrated that the purified S-protein here had the biological activity.
Using codon-optimized gene constructs to express the corresponding protein in host cells is a common strategy for increasing expression levels. It was reported that codon-optimized construct can express S-protein in mammalian cells and a reasonably high-level expression is obtained[11,17]. In this paper, we used adenovirus vector to express S-protein with codon-optimized gene and very high-level expression was observed in HEK293 cells. However, we did not successfully express S-protein in 293T cells with wild-type gene construct at a detectable level since the majority of wild-type S-protein codons were deviated from main codons of humans or animals.
Another strategy to increase expression level of S-protein with wild-type S-protein gene in mammalian cells is co-infection of vaccine virus expressing T7 polymerase[16] which facilitates the corresponding mRNA synthesis in cells when the gene clones downstream T7 promoter of the eukaryotic expression vector.
Studies have been conducted to express full-length or truncated S-protein genes in prokaryotic cells[19] and eukaryotic cells[6,16]. Expression of S-protein in E. coli leads to the accumulation of recombinant protein in inclusion bodies which requires solubilization followed by in vitro refolding. Moreover, the expressed S-proteins tend to lose some biological activities because the proteins are not modified by glycosylation which involves protein-protein interactions and receptor binding. Mammalian cells are widely used to prepare S-protein for the investigation of S-protein interaction with receptors. Although the expressed S-protein is glycosylated and functionally active using mammalian expression system, the expression level of S-protein is relatively lower than expected and the procedure involved in the expression and purification is relatively time consuming and labor intensive. We report here an adenovirus-mediated expression system for SARS S-protein. Once the recombinant adenovirus is made, the high yields of S-protein expression can be obtained in a shorter time period, thus making the procedure more time- and cost-effective.
Many adenovirus expression systems are commercially available and can be used to exploit the different strategies to construct recombinant adenoviruses. The Adeno-XTM expression system we used in this paper could exploit the ligation method to construct recombinant adenovirus, which makes the recombination more efficient. The new MCS sequence of pShuttle vector modified in this paper makes it more flexible for cloning.
Since the replication-deficient recombinant adenovirus grows and propagates extremely fast in HEK293 cells, the conditions for S-protein expression in these cells should be accurately controlled and optimized. In general, the high-titer virus used and the long expression time maintained result in a high level of expression. Nevertheless, exposure of the cells to the high-titer adenovirus for a long time lends to decrease cell viability, in which intracellular components of the dead cells are released into media, interfering with the following S-protein purification.
The authors thank Dr. Lu Jinhua (National University of Singapore Medical School) for thoughtful scientific discussion and support pertaining to this work as well as Dr. Micheal Farzan (Brigham and Women’s Hospital, Harvard University) for kindly providing the codon-optimized S-protein gene sequence.
S- Editor Wang J L- Editor Wang XL E- Editor Ma WH
| 1. | Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3147] [Cited by in RCA: 3072] [Article Influence: 139.6] [Reference Citation Analysis (0)] |
| 2. | Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88-S97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 740] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 3. | Drosten C, Preiser W, Günther S, Schmitz H, Doerr HW. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med. 2003;9:325-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 4. | Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1538] [Cited by in RCA: 1559] [Article Influence: 70.9] [Reference Citation Analysis (0)] |
| 5. | Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen MH. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1864] [Cited by in RCA: 1882] [Article Influence: 85.5] [Reference Citation Analysis (0)] |
| 6. | Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4113] [Cited by in RCA: 4603] [Article Influence: 209.2] [Reference Citation Analysis (0)] |
| 7. | To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol. 2004;203:740-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 8. | Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3643] [Cited by in RCA: 4149] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 9. | Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, Subbarao K, Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci U S A. 2004;101:6641-6646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 341] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 10. | Zeng F, Chow KY, Hon CC, Law KM, Yip CW, Chan KH, Peiris JS, Leung FC. Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. Biochem Biophys Res Commun. 2004;315:1134-1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 526] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 12. | Bukreyev A, Lamirande EW, Buchholz UJ, Vogel LN, Elkins WR, St Claire M, Murphy BR, Subbarao K, Collins PL. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122-2127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 206] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 13. | Mizuguchi H, Kay MA. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum Gene Ther. 1998;9:2577-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 268] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 14. | Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3984] [Cited by in RCA: 4349] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 15. | Krauspe R, Scheer A. Coomassie brilliant blue G-250 dye-binding technique for determination of autolytic protein breakdown in Euglena gracilis and comparison to other methods of autolysis measurement. Anal Biochem. 1986;153:242-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Biophys Res Commun. 2003;312:1159-1164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 301] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 17. | Babcock GJ, Esshaki DJ, Thomas WD, Ambrosino DM. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552-4560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 186] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 18. | Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197-3201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 538] [Cited by in RCA: 551] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 19. | Zhao JC, Zhao ZD, Wang W, Gao XM. Prokaryotic expression, refolding, and purification of fragment 450-650 of the spike protein of SARS-coronavirus. Protein Expr Purif. 2005;39:169-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |