Published online Oct 7, 2006. doi: 10.3748/wjg.v12.i37.5978
Revised: March 12, 2005
Accepted: February 25, 2006
Published online: October 7, 2006
AIM: To investigate whether the stimulation of peripheral blood mononuclear cells (PBMNC) with the cell debris and cell extraction of different probiotic strains is similar or species specific.
METHODS: Three strains of bifidobacteria, 4 strains of lactobacilli, and E. coli nissle were sonicated and centrifuged in order to divide them into cell extract and cell debris. PBMNC were separated by density gradient and incubated for 36 h with either the cell debris or the cell extract of single strains of probiotic bacteria in doses from 102 to 108 CFU/mL. Cell supernatants were taken and interleukin (IL)-10, IL-1β, and tumor necosis factor (TNF)-α were determined by ELISA.
RESULTS: Depending on the species super-family, the strains had different stimulation patterns. Except for both L. casei strains, the cell extract of bifidobacteria and lactobacilli had less stimulating capacity than cell debris, whereas the cell extract of E. coli nissle had similar stimulating properties to that of the cell debris of the strain and significantly more stimulating capacity than that of bifidobacteria and lactobacilli. The cell debris of bifidobacteria stimulated more cytokine release than the cell debris of lactobacilli. The cell debris of lactobacilli did not have a stimulating capacity when lower concentrations were used. Neither cell extraction nor cell debris had an inhibitory effect on the production of the tested cytokines by stimulated PBMNC.
CONCLUSION: The incubation of probiotic strains, which have been used in clinical trials for inflammatory diseases, with immunocompetent cells leads to different species specific reactions. High IL-10 response to cell debris of bifidobacteria and E. coli nissle can be found. This corresponds to positive effects of bifidobacteria and E. coli nissle in clinical trials for inflammatory bowel disease compared to negative outcomes obtained with lactobacilli.
- Citation: Helwig U, Lammers KM, Rizzello F, Brigidi P, Rohleder V, Caramelli E, Gionchetti P, Schrezenmeir J, Foelsch UR, Schreiber S, Campieri M. Lactobacilli, bifidobacteria and E. coli nissle induce pro- and anti-inflammatory cytokines in peripheral blood mononuclear cells. World J Gastroenterol 2006; 12(37): 5978-5986
- URL: https://www.wjgnet.com/1007-9327/full/v12/i37/5978.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i37.5978
There is no doubt that the relationship between intestinal microflora and immune system is complex. The host has to distinguish between pathogenic bacteria and harmless commensal and must react in an adequate and not self-destructive manner[1]. There is an increasing body of evidence that chronic intestinal inflammations such as inflammatory bowel disease (IBD) are due to a disturbed relationship within the host’s immune response to the enteric microflora[2-4]. Based on these proposals the relationship between micro-flora and intestinal immune response has been intensively studied by manipulation of the enteric micro-flora with probiotic bacteria[5-11]. These are by definition “A preparation of or a product containing viable, defined products in sufficient numbers which alter the microflora (by implantation or colonization) in a compartment of the host and by exerting beneficial health effects in the host”[12]. The efficacy of E. coli nissle in therapy for ulcerative colitis[5-6] has been shown and our own clinical experience has been focused on a highly concentrated probiotic preparation (VSL#3) in preventing pouchitis, an unspecific inflammation of an ileal pouch anal anastomosis after colectomy for ulcerative colitis[9-10]. Lactobacillus GG instead has no influence on clinical outcome in Crohn’s disease[7-8]. Besides clinical findings, different studies have also shown the influence of probiotic bacteria on the local and systemic immune response in experimental colitis[13-15]. The mechanisms underlying this effect are still under investigation. One hypothesis is based on modulating pro- and anti-inflammatory cytokines[16-17]. Pro-inflammatory cytokines such as interleukin (IL)-1β and tumor necosis factor (TNF)-α play an important role in gut inflammation[17]. In Crohn’s disease and ulcerative colitis these cytokines are elevated in local inflammation area and peripheral blood cells[18-19], whereas the anti-inflammatory cytokine IL-10 is decreased in patients suffering from IBD[20]. IL-10 produced by gene manipulated bacteria, reduces toxic colitis that is associated with pro-inflammatory cytokines[21-22]. Further arguments have been found in the absence of IL-10. IL-10 deficient (KO) mice do not show any symptoms of intestinal inflammation as long as they are kept in sterile conditions, but spontaneously develop chronic colitis with a histological distribution similar to that found in Crohn’s disease after termination of the sterile conditions[23]. If the mice are fed with different lactic acid bacteria before finishing the sterile conditions, this chronic inflammation can be prevented[24]. Recently, during preventive therapy for chronic pouchitis using different probiotics as mentioned above, we investigated the cytokine tissue levels of patients with pouchitis and after induction of remission and during the following probiotic application. It is interesting to find that cytokine tissue levels of IL-10 increase during the application of the probiotic preparation, whereas anti-inflammatory cytokine-levels such as TNF-α and IL-1 remain low after the application[25].
The aim of our study was to set up an in vitro model to compare the immunomodulatory effects of different probiotic strains that have previously been evaluated in different clinical trials. For this purpose we used peripheral blood mononuclear cells (PBMNC), which are a combination of different immunogenic cells.
Blood samples were taken from 12 healthy blood donors (7 females, mean age: 44 years; 5 males, mean age: 52 years). Blood from the same donor was used for each co-incubation with all tested bacteria in different concentrations. Incubation experiments were repeated 4 to 6 times with blood from different donors. The study was performed in accordance with the Declaration of Helsinki and the local ethics committee.
PBMNC from healthy donors were separated according to Boyum[26]. Briefly, peripheral blood diluted with Hank’s balanced salt solution (HBSS) (without Ca2+ or Mg2+) (GIBCO, Karlsruhe, Germany) containing 100 U/mL heparin, was layered over a ficoll (Lymphoprep, Progen, Biotechnik, Heidelberg, Germany; specific gravity: 1.077) and centrifuged for 40 min at 400 r/min without using a frame. Cells harvested from the interface were washed in HBSS (without Ca2+ or Mg2+) and centrifuged for 10 min at 400 r/min. Supernatant was discarded and the pellet was resuspended in HBSS (without Ca2+ or Mg2+), which was repeated four times. Finally, resuspension was performed in RPMI 1640 (GIBCO, Karlsruhe, Germany) with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 50 μg/mL gentamycin (all Sigma, St. Louis, MO). Viability of cells was tested by trypan-blue, which was more than 97%. After calculation of the cells per volume, the cell count was adjusted to 500 000 cells per well and per mL.
The bacteria species and strains used in this study are listed in Table 1. Strains in bold type originated from the pharmaceutical probiotic VSL#3 (Sigma Tau, Promezia, Italy). Bifidobacterium and Lactobacillus strains were grown in MRS broth (Difco, Detroit, MI) with the addition of 0.05% L-cysteine hydrochloride monohydrate (Merck, Darmstadt, Germany). All strains were incubated anaerobically at 37°C. Mid log cultures counted by plating technique on the above mentioned media, were collected by centrifugation (8 000 × g for 3 min), washed and resuspended in 5 mL RPMI 1640 Medium (GIBCO, Karlsruhe, Germany). The bacterial suspensions were subsequently sonicated (Branson Sonifier W-250, Heinemann, Schwäbisch, Germany) at power levels 5-6 at 30% duty for 5 min to destroy cellular membranes. The sonicated suspension was centrifuged at 8 000 × g for 30 min to separate cell debris from crude cell extract. After centrifugation the supernatant containing the cell extract and the pellet containing the cell debris were taken for further investigation.
| Bifidobacterium breve: Y 8 |
| Bifidobacterium infantis: Y 1 |
| Bifidobacterium longum: Y 10 |
| E. coli: Stamm Nissle 1917 (Mutaflor, Ardeypharm, Herdecke, Germany) |
| Lactobacillus azidophilus: MB 443 |
| Lactobacillus casei subspecies rhamnosus: Lactobacillus GG (LGG, Valio,Helsinki, Finland): ATCC 53103 |
| Lactobacillus casei: MB 451 |
| Lactobacillus delbrueckii subspecies bulgaricus: MB 453 |
| Lactobacillus plantarum: MB 452 |
Bacterial cell debris and extract were applied at concentrations ranging from 1 × 103 to 1 × 1010 colony forming units (CFUs)/mL. One hundred μL at specific concentration was transferred to 900 μL medium containing 500 000 mononuclear cells and co-incubated at 37°C and 50 mL/L CO2 for 36 h. Supernatants were collected and stored at -20°C until assay. The viability of PBMNC was checked by the trypan blue test. On each incubation plate a positive control with LPS added to PBMNC and a negative control without stimulus were investigated. The same set-up was used in order to examine the ability of probiotics to inhibit LPS-induced cytokine release. PBMNC were incubated with bacterial cell extract and debris for 10 min, then LPS was added in a concentration of 100 ng/μL.
Cytokine quantification in culture supernatant was analysed by commercially available sandwich enzyme linked immunosorbent assay (ELISA).
TNF-α was detected using anti-human TNF-α monoclonal “capture” antibody (MAB 610, R&D Systems, Minneapolis, MN, USA) and biotinylated “detection” antibody (BAF 210 R&D Systems, Minneapolis, MN, USA) with o-phenylenediamine buffer/H2O2 (Sigma, Steinheim, Germany) as substrate. Standard procedure was performed by using recombinant human TNF-α (210-TA, R&D Systems, Minneapolis, MN, USA). The absorbance values of the sample were read at 490 nm on an ELISA plate reader. ELISA measurements were performed in duplicates or triplicates.
IL-1β was detected using anti-human IL-1β monoclonal “capture” antibody (MAB 601, R&D Systems, Minneapolis, MN, USA) and biotinylated “detection” antibody (BAF 201 R&D Systems, Minneapolis, MN, USA) with o-phenylenediamine buffer/H2O2 (Sigma, Steinheim, Germany) as substrate. Standard procedure was performed by using recombinant human IL-1β (201-LB, R&D Systems, Minneapolis, MN, USA). The absorbance values of the sample were read at 490 nm on an ELISA plate reader. ELISA measurements were performed in duplicates or triplicates.
IL-10 was detected using anti-human IL-10 “capture” antibody (18551A Pharmingen, San Diego, CA, USA) and biotinylated “detection” antibody (18562D, Pharmingen, San Diego, CA, USA) with o-phenylenediamine buffer/H2O2 (Sigma, Steinheim, Germany) as substrate. Standard procedure was performed by using recombinant human IL-10 (19701V, Pharmingen, San Diego, CA, USA, USA). The absorbance values of the sample were read at 490 nm on an ELISA plate reader. ELISA measurements were performed in duplicates or triplicates.
Data of cytokine concentration were presented as mean ± SE and expressed in pg/mL. For quantification of stimulating capacity of bacteria at different concentrations, cytokine concentration was resumed as an area under the curve (AUC) and described as (mean ± SE) AUC. Statistical significance was calculated by the Mann-Whitney-Rank test and expressed as P-value.
Results from cytokine production in PBMNC after stimulation with LPS (100 ng/mL) were pooled (144 samples from 12 different donors). The mean cytokine production in stimulated PBMNC was 186.5 ± 125.6 pg/mL for IL-10, 1875.6 ± 1381.2 pg/mL for IL-1β and 356.0 ± 249.1 pg/mL for TNF-α. Cytokine production in non-stimulated PBMNC was under detection limit of the ELISA.
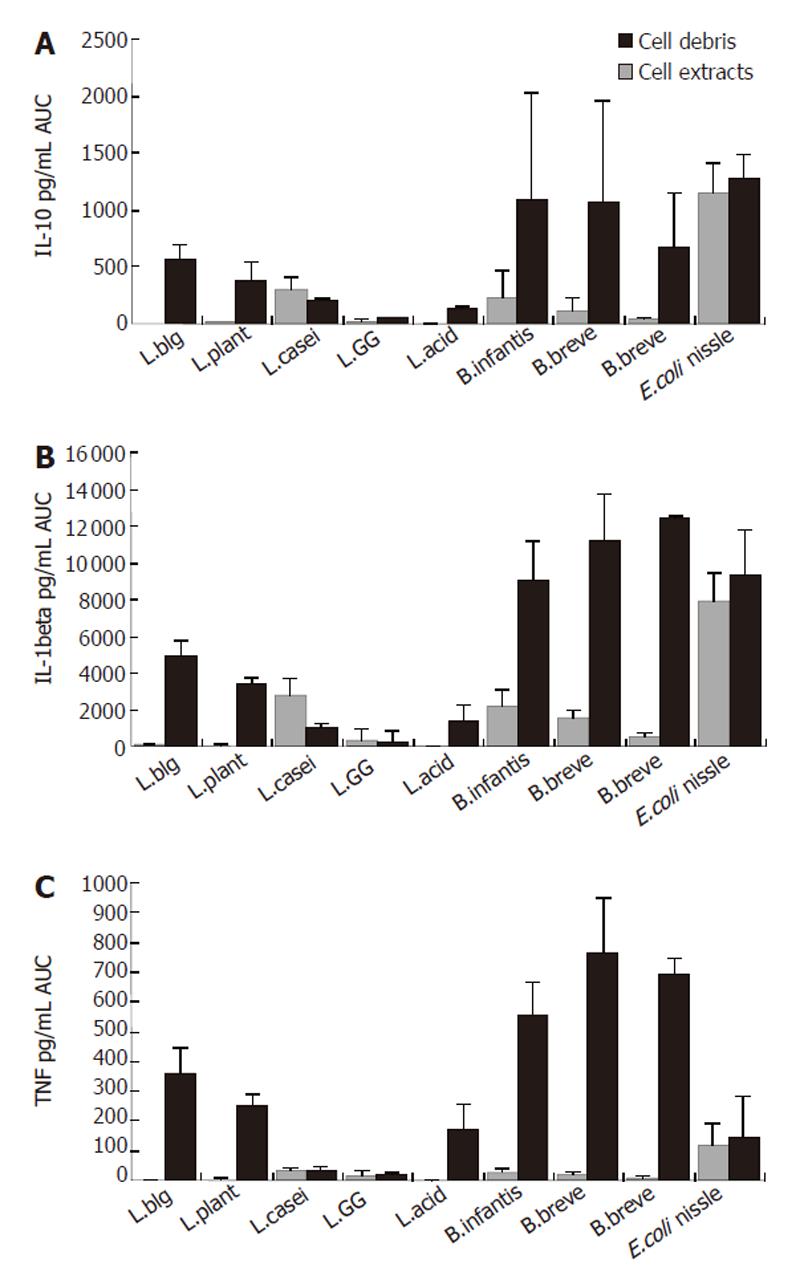
Generally, cytokine production in PBMNC induced by bacteria differed in cell debris and extract between bacteria and depended on the applied concentration used (Figure 1).
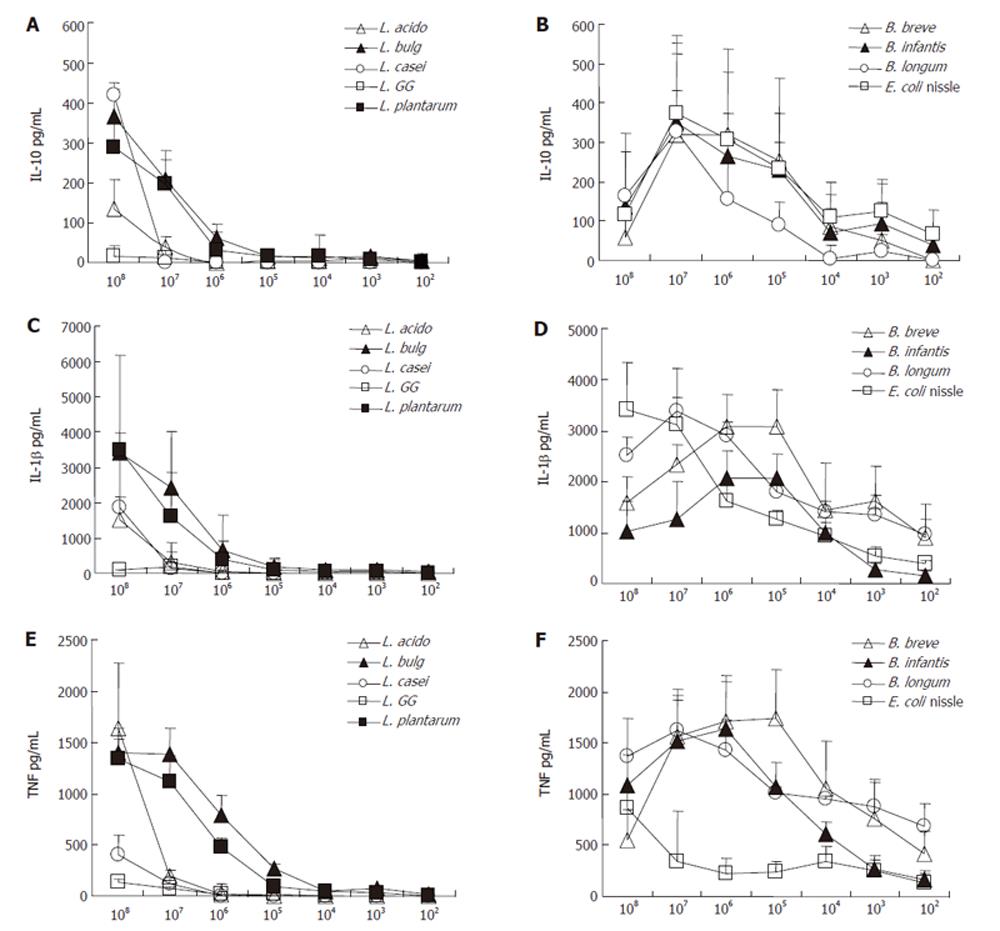
Lactobacilli: Cell extracts from all applied lactobacilli induced IL-10 concentration only weakly, whereas no difference was found between species (data not shown). The stimulation by cell debris of these strains led to higher concentrations of IL-10 whereas significance only reached in L. azidophilus MB443, L. delbrueckii subsp. bulgaricus MB453 and L. plantarum MB452 (AUC: L. azidophilus MB443: 1.0 ± 1.5 pg/mL; L. delbrueckii subsp. bulgaricus MB453: 3.8 ± 4.65 pg/mL; L. plantarum MB452: 9.9 ± 13.8 pg/mL). The stimulation by cell debris of both L. casei subs. (MB 451 and L. GG)) did not differ from that of cell extract of these strains. As shown in Figures 2 and 3, the cell debris of lactobacilli had a weak stimulation capacity at concentrations less than 105 CFU/mL.
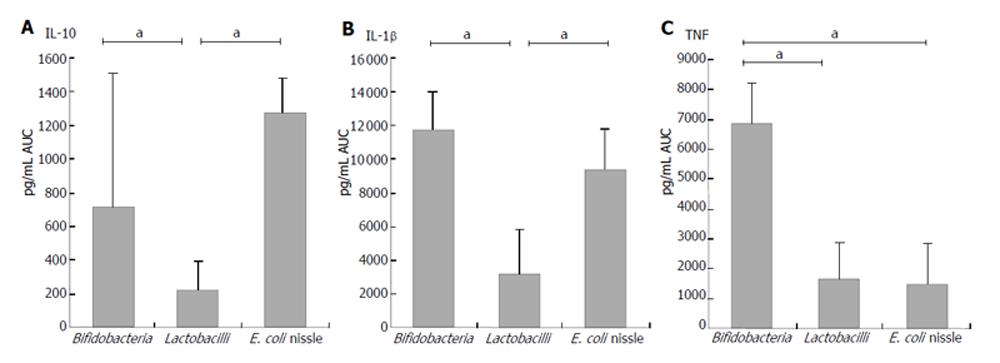
Bifidobacteria: Cell debris of each bifidobacteria strain stimulated IL-10 production in PBMNC more significantly than their cell extracts (AUC: B. breve Y8 cell debris: 1062.7± 889.9 pg/mL; cell extract: 182.6 ± 177.5 pg/mL; AUC B. longum Y10 cell debris: 682.7 ± 466.9 pg/mL; cell extract: 30.5 ± 28.9 pg/mL; AUC B. infantis Y1 cell debris: 1095.6± 925.3 pg/mL; cell extract: 228.3 ± 233.0 pg/mL; P < 0.05) (Figure1). No difference was found between different bifidobacteria species when the stimulation capacity of cell extracts or cell debris was compared. Cell debris from bifidobacteria stimulated IL-10 production in PBMNC more significantly than lactobacilli (AUC cell debris of bifidobacteria: 710.1 ± 795.5 pg/mL; AUC cell debris of lactobacilli: 219.6 ± 174.7 pg/mL; P < 0.02) (Figure 4).
As shown in Figure 2 and Figure 3, the cell debris of bifidobacteria had a weak stimulating capacity at the concentration lower than 103 CFU/mL. Moreover, the highest concentration (108 CFU/mL) of bifidobacteria had a less stimulating capacity than the lower concentration (107 CFU/mL), whereas the viability measured by trypan blue test was over 97%.
E. coli nissle: Cell extracts and debris of E. coli nissle led to similar IL-10 concentrations (AUC cell debris: 1270.7 ± 210.9 pg/mL; cell extract: 1154.8 ± 264.0 pg/mL) (Figure 1). The stimulation ability of cell extract to produce IL-1β in PBMNC was significantly higher in E. coli nissle than in lactobacilli or bifidobacteria (P < 0.05). The cell debris of E. coli nissle had a similar stimulating capacity to bifidobacteria but a more significant capacity than lactobacilli (P < 0.02) (Figure 4). The cell debris and extract of E. coli nissle even had a stimulating capacity at low concentrations (Figure 3). The highest concentration (108 CFU/mL) of E. coli nissle had a less stimulating capacity than the lower concentration (107 CFU/mL), whereas the viability measured by trypan blue test was over 97%.
IL-1β concentration
Lactobacilli: As shown in Figure 1 cell extracts from all applied lactobacilli induced IL-1β concentration only weakly, whereas no difference was found between species (data not shown). The stimulation by cell debris of L. acidophilus, L. delbrueckii subsp. bulgaricus and L. plantarum led to higher concentrations of IL-1β (AUC: L. azidophilus MB443: 1282.2 ± 987.9pg/mL; L. delbrueckii subsp. bulgaricus MB453: 4881.5 ± 893.8 pg/mL; L. plantarum MB452: 3390.8 ± 288.9 pg/mL) (P < 0.05) (Figure 1). The stimulation by cell debris did not differ from cell extract of L. casei subs. rhamnosus (L.GG) and L. casei MB451. As shown in Figures 2 and 3 , the cell debris of lactobacilli had only a weak stimulating capacity at concentrations of less than 106 CFU/mL.
Bifidobacteria: Cell debris of each bifidobacteria strain stimulated IL-1β production in PBMNC more significantly than their cell extract (AUC: B. breve Y8 cell debris: 11 152.9 ± 2547.7 pg/mL; cell extract: 1488.3 ± 454.0 pg/mL; AUC B. longum Y10 cell debris: 12 364.0 ± 192.5 pg/mL; cell extract: 491.0 ± 190.6 pg/mL; AUC B. infantis Y1 cell debris: 9018.8 ± 2190 pg/mL; cell extract: 2142.3 ± 925.0 pg/mL; P < 0.05) (Figure 1). No difference was found between bifidobacteria species when the stimulating capacity of cell extracts or cell debris was compared. The cell debris from bifidobacteria led to higher IL-1β concentrations in supernatant of PBMNC than cell debris of lactobacilli (AUC cell debris of bifidobacteria: 11692.8 ± 2283.2 pg/mL; AUC cell debris of lactobacilli: 3143.3 ± 2689.0 pg/mL; P < 0.02) (Figure 4). As shown in Figure 2, the cell debris of B. breve Y8 and B. longum Y10 even had a stimulating capacity at low concentrations (103 CFU/mL and 102 CFU/mL), whereas the IL-1β production in PBMNC was weak when incubated with B. infantis Y1 at a concentration of less than 104 CFU/mL. The highest cell debris concentration (108 CFU/mL) of bifidobacteria had a less stimulating ability to produce IL-1β in PBMNC than its lower concentration, whereas the viability measured by trypan blue test was over 97%.
E. coli nissle: Cell extracts and debris of E. coli nissle led to similar IL-1β concentrations (AUC cell debris: 9334.8 ± 2486.1 pg/mL; cell extract: 7875.0 ± 1595.3 pg/mL) (Figure 1). The stimulating ability of the cell extract of E. coli nissle to produce IL-1β in PBMNC was significantly higher than that of lactobacilli or bifidobacteria (P < 0.05). The cell debris of E. coli nissle had a similar stimulating capacity to bifidobacteria but a more significant capacity than lactobacilli (P < 0.02) (Figure 4). The cell debris and extract of E. coli nissle even had a stimulating capacity at low concentrations (Figure 3). The highest concentration of debris of E. coli nissle had a less stimulatory capacity than lower concentration.
Lactobacilli: Cell extracts from L. azidophilus MB443, L. delbrueckii subsp. bulgaricus MB453, and L. plantarum MB452 induced TNF-α concentration only weakly. The stimulation by cell debris of these strains led to higher TNF-α production (AUC: L. azidophilus MB443: 1695.3 ± 879.3 pg/mL; L. delbrueckii subsp. bulgaricus MB453: 3593.2 ± 822.1 pg/mL; L. plantarum MB452: 2466.5 L ± 433.3 pg/mL) (P < 0.05) (Figure 1). The stimulation by cell debris did not differ from cell extract of L. casei subs. rhamnosus (L.GG) and L. casei MB451. As shown in Figures 2 and 3, the cell debris of lactobacilli only had a weak stimulating capacity at concentrations of less than 107 CFU/mL.
Bifidobacteria: Cell debris of each bifidobacteria strain led to higher TNFα concentrations in supernatant of PBMNC than their cell extract (AUC: B. breve Y8 cell debris: 7645.5 ± 1823.1 pg/mL; cell extract: 180.1 ± 85.5 pg/mL; AUC B. longum Y10 cell debris: 6951.2 ± 522.7 pg/mL; cell extract: 83.6 ± 40.0 pg/mL; AUC B. infantis Y1 cell debris: 5584.8 ± 1098.8 pg/mL; cell extract: 290.5 ± 103.6 pg/mL) (P < 0.05). No difference was found between bifidobacteria species when the stimulating capacity of cell extract or cell debris was compared. The cell debris from bifidobacteria led to higher TNFα concentrations in supernatant of PBMNC than cell debris from lactobacilli or E. coli nissle (AUC cell debris of bifidobacteria: 6882.4 L ± 1355.0 pg/mL; AUC cell debris of lactobacilli: 1630.8 ± 1265.1 pg/mL; AUC cell debris of E. coli nissle: 1466.6 ± 1356.0 pg/mL) (P < 0.02) (Figure 4). As shown in Figure 2, the cell debris from B. breve Y8 and B. longum Y10 even had a stimulating capacity at low concentrations (103 CFU/mL and 102 CFU/mL), whereas the TNF-α production in PBMNC was weak when incubated with B. infantis Y1 at a concentration of less than 104 CFU/mL. The highest concentration (108 CFU/mL) of bifidobacteria had no strong stimulating ability to produce TNF-α in PBMNC, whereas the viability measured by trypan blue test was over 97%.
E. coli nissle: High concentrations (108 CFU/mL) of cell debris of E. coli nissle led to high concentrations of TNF-α, whereas lower concentrations of cell debris and extracts led to lower concentrations of TNF-α (Figure 3) (AUC cell debris: 1466.6 ± 1356 pg/mL; cell extract: 1153.0 ± 748.4 pg/mL) The cell debris of E. coli nissle had a similar stimulating capacity similar to lactobacilli but a significantly less capacity than that of bifidobacteria (P < 0.02) (Figure 4).
None of the bacterial cell debris and extract of lactobacilli, bifidobacteria or E. coli nissle had an inhibitory effect on the cytokine production in PBMNC. Pre-incubation with cell debris or with cell extract and stimulation with LPS led to similar cytokine production of LPS alone or cell debris alone (data not shown).
Different probiotic strains used in clinical trials have shown prophylactic properties in different inflammatory diseases of the gastrointestinal tract, such as Crohn’s disease, ulcerative colitis, pouchitis, antibiotic-associated colitis and traveller’s diarrhoea[27-28]. Recently, we have shown that the IL-10 concentration in the mucosa of ileo- anal pouch tissue is elevated after administration of probiotics[25]. In this in vivo approach we used a highly concentrated probiotic preparation containing different lactic acid bacteria. Our hypothesis is that a high concentration of bacteria contributes to these clinical results and immunologic findings[3]. However, the specific property of different strains remains unclear. Therefore, we investigated an in vitro model in order to test different probiotic strains and species, which are used in clinical practice to prevent inflammatory diseases, in order to understand the pro- and anti-inflammatory properties of specific strains. There are several studies on the induction of cytokines by cell components of lactic acid bacteria to induce cytokines[29-31]. However, systematic analysis of probiotic bacteria used in clinical practice for the prevention of inflammatory disease, has not been performed until now in a human cell model. For this purpose we used PBMNC which are easily available and express toll-like-receptor (TLR) 2 and TLR 4 as well as CD14 which are shown to mediate immune response to microbial components as peptidoglycan and lipoteichoic acid[32-33]. Until now there is no report on comparison of dose response over a broad range of different concentrations of probiotic bacteria used in clinical practice for prevention of inflammatory bowel disease. Our findings on PBMNC indicate that stimulation by lactobacilli works in a dose-dependent way. High doses of cell debris could stimulate PBMNC to produce pro- and anti-inflammatory cytokines. The cell extract has a less stimulating capacity in a dose-dependent manner. An interesting finding is that the cell debris of L. delbrueckii subsp. bulgaricus MB453 and L. plantarum MB452 stimulates PBMNC when used at concentrations higher than 104 CFU/mL, while both L. azidophilus MB443 and L. casei MB451 strains only require concentrations higher than 106 and 105 CFU/mL. Cell debris of L. casei supsp. rhamnosus (L.GG) had a very low stimulating capacity compared to other strains (Figure 1). The weak or even absent reaction at high concentrations of lactobacilli (106 or 105 CFU/mL) is not suspected. This phenomenon is reproducible in different blood donors and exists in all different lactobacilli strains when used for this examination. Schultz and co-workers[34] recently showed that L. casei subsp. rhamnosus (L.GG), which had the lowest stimulating capacity in our study, induces immunologic tolerance to granulocytes after oral administration for several weeks. Since lactobacillus strains normally are early inhabitants of the human gastrointestinal tract, oral tolerance to low concentrations of lactobacillus strains might generally develop[35]. Although this is expected for bifidobacteria, they stimulate pro- and anti-inflammatory cytokines more significantly than lactobacilli. But the stimulation pattern is different. The highest concentration of bifidobacteria induces PBMNC to produce less pro- and anti-inflammatory cytokines than lower concentrations of the strain. Whether the lower induction of cytokine release in incubation with highly concentrated cell debris is due to deletion or apoptosis of PBMNC remains unclear. Toxic reaction or a reaction resulting in direct cell death can be excluded by the trypan blue control test which provides information about the functionality of cell membrane but not about the metabolic condition of cells. Recently, it has been proposed in a different model that bifidobacteria strains induce oral tolerance[36] but also induction of oral tolerance to
E. coli and lactobacilli has been reported[37-38]. The stimulating capacity of E. coli nissle shows a different pattern. The cell extract and debris of E. coli nissle have a similar ability to produce cytokines. Interestingly, the cell debris and extract of E. coli nissle at low concentrations can stimulate epithelial HT29/19 cells to produce the chemotactic factor interleukin-8 (IL-8), whereas the cell debris and extract of lactobacilli and bifidobacteria do not stimulate epithelial HT29/19 cells to release IL-8[38]. E. coli nissle, which has been shown to be effective in maintaining remission of ulcerative colitis, has a high stimulating capacity for IL-10 and IL-1β compared to other strains, but a low capacity for TNF-α. Bifidobacteria of the probiotic composition VSL#3 of bifidobacteria, which can prevent inflammatory bowel disease, can stimulate PBMNC to produce IL-10[9,10]. L. GG can weakly stimulate PBMNC to produce IL-10 and has no positive effect on inflammatory bowel disease[7-8]. This is consistent with the findings in another study[25]. L. GG has been primarily used in trials for prevention of relapses in Crohn’s disease. It has been recently reported that Crohn’s disease is associated with the polymorphism of the nucleotide-binding oligomerization domain 2 (NOD 2)[39-40]. NOD 2 is a regulator of TLR 2-mediated response to microbial agents[41] and Gram-positive bacteria like lactobacilli are typical ligands for TLR 2[32]. Since the function of mutations in the NOD 2 gene in Crohn’s disease is not clear[42], explanation about the lacking effect of probiotics on Crohn’s disease is warranted.
In conclusion, the ability of probiotic bacteria to stimulate PBMNC is different. Compared to E. coli nissle and bifidobacteria, lactobacilli debris exerts effects only at high concentrations. Whereas the extract of lactobacilli and bifidobacteria has only weak effects, while the cell extract and debris of E. coli nissle have similar effects. The higher IL-10 response to E. coli nissle and bifidobacteria corresponds to the positive effect of these probiotic strains on inflammatory bowel disease compared to negative outcomes obtained with lactobacilli.
S- Editor Wang J L- Editor Wang XL E- Editor Bi L
| 1. | MacDonald TT, Pettersson S. Bacterial regulation of intestinal immune responses. Inflamm Bowel Dis. 2000;6:116-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Sartor RB. Review article: Role of the enteric microflora in the pathogenesis of intestinal inflammation and arthritis. Aliment Pharmacol Ther. 1997;11 Suppl 3:17-22; discussion 22-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Campieri M, Gionchetti P. Bacteria as the cause of ulcerative colitis. Gut. 2001;48:132-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 144] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Janowitz HD, Croen EC, Sachar DB. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm Bowel Dis. 1998;4:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet. 1999;354:635-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 700] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 6. | Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, Stolte M. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 1997;11:853-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 478] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 7. | Prantera C, Scribano ML, Falasco G, Andreoli A, Luzi C. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn's disease: a randomised controlled trial with Lactobacillus GG. Gut. 2002;51:405-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 363] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 8. | Schultz M, Timmer A, Herfarth HH, Sartor RB, Vanderhoof JA, Rath HC. Lactobacillus GG in inducing and maintaining remission of Crohn's disease. BMC Gastroenterol. 2004;4:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 215] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 9. | Gionchetti P, Rizzello F, Venturi A, Brigidi P, Matteuzzi D, Bazzocchi G, Poggioli G, Miglioli M, Campieri M. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1077] [Cited by in RCA: 956] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 10. | Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108-114. [RCA] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 611] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 11. | Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 708] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 12. | Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics--approaching a definition. Am J Clin Nutr. 2001;73:361S-364S. [PubMed] |
| 13. | Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 245] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 14. | Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 481] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 15. | Fukushima K, Sasaki I, Ogawa H, Naito H, Funayama Y, Matsuno S. Colonization of microflora in mice: mucosal defense against luminal bacteria. J Gastroenterol. 1999;34:54-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620-1633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 726] [Cited by in RCA: 718] [Article Influence: 34.2] [Reference Citation Analysis (1)] |
| 17. | Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1493] [Cited by in RCA: 1484] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 18. | Schreiber S, MacDermott RP, Raedler A, Pinnau R, Bertovich MJ, Nash GS. Increased activation of isolated intestinal lamina propria mononuclear cells in inflammatory bowel disease. Gastroenterology. 1991;101:1020-1030. [PubMed] |
| 19. | Reinecker HC, Steffen M, Witthoeft T, Pflueger I, Schreiber S, MacDermott RP, Raedler A. Enhanced secretion of tumour necrosis factor-alpha, IL-6, and IL-1 beta by isolated lamina propria mononuclear cells from patients with ulcerative colitis and Crohn's disease. Clin Exp Immunol. 1993;94:174-181. [RCA] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 648] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 20. | Schreiber S, Heinig T, Thiele HG, Raedler A. Immunoregulatory role of interleukin 10 in patients with inflammatory bowel disease. Gastroenterology. 1995;108:1434-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 260] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 21. | Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1049] [Cited by in RCA: 1012] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 22. | Fiorucci S, Distrutti E, Mencarelli A, Barbanti M, Palazzini E, Morelli A. Inhibition of intestinal bacterial translocation with rifaximin modulates lamina propria monocytic cells reactivity and protects against inflammation in a rodent model of colitis. Digestion. 2002;66:246-256. [RCA] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3189] [Cited by in RCA: 3225] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 24. | Madsen KL, Doyle JS, Tavernini MM, Jewell LD, Rennie RP, Fedorak RN. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 25. | Ulisse S, Gionchetti P, D'Alò S, Russo FP, Pesce I, Ricci G, Rizzello F, Helwig U, Cifone MG, Campieri M. Expression of cytokines, inducible nitric oxide synthase, and matrix metalloproteinases in pouchitis: effects of probiotic treatment. Am J Gastroenterol. 2001;96:2691-2699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 133] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Böyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl. 1968;97:77-89. [PubMed] |
| 27. | Gionchetti P, Rizzello F, Venturi A, Campieri M. Probiotics in infective diarrhoea and inflammatory bowel diseases. J Gastroenterol Hepatol. 2000;15:489-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Marteau P, Seksik P, Jian R. Probiotics and intestinal health effects: a clinical perspective. Br J Nutr. 2002;88 Suppl 1:S51-S57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 85] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Miettinen M, Matikainen S, Vuopio-Varkila J, Pirhonen J, Varkila K, Kurimoto M, Julkunen I. Lactobacilli and streptococci induce interleukin-12 (IL-12), IL-18, and gamma interferon production in human peripheral blood mononuclear cells. Infect Immun. 1998;66:6058-6062. [PubMed] |
| 30. | Miettinen M, Vuopio-Varkila J, Varkila K. Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun. 1996;64:5403-5405. [PubMed] |
| 31. | Miettinen M, Lehtonen A, Julkunen I, Matikainen S. Lactobacilli and Streptococci activate NF-kappa B and STAT signaling pathways in human macrophages. J Immunol. 2000;164:3733-3740. [PubMed] |
| 32. | O'Neill LA. TLRs: Professor Mechnikov, sit on your hat. Trends Immunol. 2004;25:687-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5:971-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 594] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 34. | Schultz M, Linde HJ, Lehn N, Zimmermann K, Grossmann J, Falk W, Scholmerich J. Immunomodulatory consequences of oral administration of Lactobacillus rhamnosus strain GG in healthy volunteers. J Dairy Res. 2003;70:165-173. [RCA] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 93] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 35. | Edwards CA, Parrett AM. Intestinal flora during the first months of life: new perspectives. Br J Nutr. 2002;88 Suppl 1:S11-S18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Duchmann R, May E, Heike M, Knolle P, Neurath M, Meyer zum Buschenfelde KH. T cell specificity and cross reactivity towards enterobacteria, bacteroides, bifidobacterium, and antigens from resident intestinal flora in humans. Gut. 1999;44:812-818. [RCA] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 189] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Tanaka K, Ishikawa H. Role of intestinal bacterial flora in oral tolerance induction. Histol Histopathol. 2004;19:907-914. [PubMed] |
| 38. | Lammers KM, Helwig U, Swennen E, Rizzello F, Venturi A, Caramelli E, Kamm MA, Brigidi P, Gionchetti P, Campieri M. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol. 2002;97:1182-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 100] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3903] [Article Influence: 162.6] [Reference Citation Analysis (0)] |
| 40. | Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925-1928. [RCA] [DOI] [Full Text] [Cited by in Crossref: 769] [Cited by in RCA: 769] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 41. | Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 603] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 42. | Girardin SE, Hugot JP, Sansonetti PJ. Lessons from Nod2 studies: towards a link between Crohn's disease and bacterial sensing. Trends Immunol. 2003;24:652-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 102] [Article Influence: 4.6] [Reference Citation Analysis (0)] |