Published online Jul 14, 2006. doi: 10.3748/wjg.v12.i26.4219
Revised: February 20, 2006
Accepted: February 28, 2006
Published online: July 14, 2006
AIM: To investigate the clinical significance and presence of mutations in the surface (S) and overlapping polymerase gene of hepatitis B patients with coexisting HBsAg and anti-HBs.
METHODS: Twenty-three patients with chronic hepatitis B were studied. Of the 23 patients, 11 were both positive for hepatitis B virus (HBV) surface antigen (HBsAg) and antibody to HBV surface antigen (anti-HBs), 12 were negative for anti-HBs while positive for HBsAg. DNA was extracted from 200 μL serum of the patients. Nucleotide of the surface and overlapping polymerase gene from HBV-infected patients was amplified by PCR, and the PCR products were sequenced.
RESULTS: Forty-one mutations were found within the surface gene protein of HBV in 15 patients (10 with coexisting HBsAg and anti-HBs). Six (14.6%) out of 41 mutations were located at “α” determinant region in 5 patients (4 positive for HBsAg and anti-HBs). Eleven mutations (26.8%) occurred in the downstream or upstream of “α” determinant region. Lamivudine (LMV)-selected mutations were found in three patients who developed anti-HBs, which occurred in amino acid positions (196, 198, 199) of the surface protein and in YMDD motif (M204I/V) of the polymerase protein simultaneously. Presence of these mutations did not relate to changes in ALT and HBV DNA levels.
CONCLUSION: Besides mutations in the “α” deter-minant region, mutations at downstream or upstream of the “α” determinant region may contribute to the development of anti-HBs. These mutations do not block the replicating competency of HBV in the presence of high titer of anti-HBs.
- Citation: Lu HY, Zeng Z, Xu XY, Zhang NL, Yu M, Gong WB. Mutations in surface and polymerase gene of chronic hepatitis B patients with coexisting HBsAg and anti-HBs. World J Gastroenterol 2006; 12(26): 4219-4223
- URL: https://www.wjgnet.com/1007-9327/full/v12/i26/4219.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i26.4219
Hepatitis B virus (HBV) infection leads to a wide spectrum of liver diseases, including acute self-limited infection, asymptomatic carrier state, fulminant and chronic hepatitis, which could result in life-threatening sequelae, such as liver cirrhosis and hepatocellular carcinoma[1]. In acute self-limited infection, clearance of HBV is associated with seroconversion from HBsAg to anti-HBs due to the coordination of humoral and cellular immune response[2,3]. However, this condition is very rare in chronic infection patients, especially in patients infected at birth, partly because of inadequate humoral and cellular immunity of the host[4]. Furthermore, the prevalence of HBV mutations that could escape from humoral and cellular immunity may result in persistent virus infection.
The S gene of HBV has three open reading frames (ORF), including preS1, preS2 and S region. The surface gene contains a neutralizing epitope named “α” determinant region located at the condon positions 124-147. Mutations in this region could alter the antigenicity of HBsAg, causing failure of anti-HBs to neutralize HBsAg and escaping from the host’s immune system, resulting in active viral replication and liver disease[5,6]. It is reported that mutations of some epitopes located at downstream of the “α” determinant region may also affect the neutralization domain[7]. The surface gene overlaps with the calatytic domains of polymerase[7]. Thus, mutations in the surface gene have an effect on the polymerase gene while the polymerase gene mutations also impact the surface gene[8,9].
The present study was to analyze the prevalence of mutations in the surface and polymerase gene of HBV in patients with coexisting anti-HBs and HBsAg.
Sera were obtained from 23 Chinese patients with chronic HBV infection. Presence of HBsAg, anti-HBs, hepatitis e antigen (HBeAg), antibody to HBeAg (anti-HBeAg) was detected by commercially available kits according to the instructions. All patients were positive for HBsAg. Of the 23 patients, 11 were positive for anti-HBs (No. 1 to 11) and 20 were positive for HBeAg. Fifteen out of the 23 patients had elevated alanine aminortransferases (ALT) levels. Virological and biochemical parameters of 8 patients positive for anti-HBs and HBsAg at the time of HBV sequence analysis were analyzed. No patients had a history of HBV vaccination or hyper immune globulin therapy. Three patients positive for HBsAg and anti-HBs had a history of lamivudine (LMV) therapy for more than 1 year. All patients were negative for antibody to hepatitis C virus. Sera were stored at -20°C for DNA extraction.
For polymerase chain reaction (PCR), primers were synthesized according to the published sequences. Sequences of the primers for amplifying the surface gene of HBV are as follows: HBV S1: 5’-TTACAGGCGGGGTTTTTC-3’ (nt 197, sense); HBV S2: 5’- AAGGGACTCAAGATG-3’-(nt 789, anti-sense). Primers of HBV P1, 5’-GTATTCCCATCCCATCATCC-3’ (nt 599, sense) and HBV P2, 5’-CAAGGCAGGATAGCCACATT-3’ (nt 1033, anti-sense) were used for amplification of polymerase gene of HBV.
DNA was extracted from 200 μL serum using a blood DNA kit (Omega, USA). Two microlitres of DNA template and 1 μL of each of the primers, 2 μL of 10 × dNTP and 0.5 U of Taq DNA polymerase (Promega, US) were used in a volume of 50 μL for PCR. After denaturation at 94°C for 5 min, the reaction for amplification of the surface gene with primers HBV S1 and HBV S2 was carried out at 94°C for 30 s, at 56°C for 1 min, and at 72°C for 1 min for 35 cycles, and a final extension at 72°C for 10 min. The reaction with primers HBV P1 and HBV P2 for amplifying the polymerase gene was performed at 94°C for 30 s, at 56°C for 30 min, at 72°C for 1 min for 35 cycles, and a final extension at 72°C for 10 min.
The PCR products were purified by centrifugation. Direct sequence of the gene was determined using Taq DyeDeoxy terminator sequencing kits. Sequencing reactions were analyzed on an automated DNA sequencer (model 377, ABI100, Applied Biosystem). Deduced amino acid sequences were compared with the reported consensus sequence of genotype C, subtype adw, HBV clone (PAK66, PIWK146). Mutations were determined as sequence different from the consensus sequence.
The nucleotide sequence data presented in this paper could be found in the DDBJ/EMBL/GeneBank nucleotide sequence databases with the access numbers AB014381, AB033554, AY812744, AY812743, AY800249, AY123424, AF100309.
Two-tailed Student’s t test was used to assess the difference in ALT levels, age, HBV DNA levels between the two groups of patients. Fisher’s exact test was used for the analysis of difference in mutations between the two groups. P < 0.05 was considered statistically significant.
Comparison of the clinical features between the two groups of patients is shown in Table 1. There was no significant difference in ALT levels, age, HBV DNA levels between the two groups (P > 0.05). The relevant biochemical and virological parameters of 8 patients (No.1 to 6, No.8 and 10) are shown in Table 2.
| Groups | |||
| Clinical factors | Positive anti-HBs | Negative anti-HBs | P |
| Age in years | 40.3 ± 13.2 | 42.5 ± 14.7 | 0.7672 |
| Sex, M/F | 5/6 | 7/5 | 0.6531 |
| Patients with HbsAg, n (%) | 11 (100) | 12 (100) | |
| Patients with HbeAg, n (%) | 9 (81.8) | 11 (91.7) | |
| Patients with anti-HBe, n (%) | 1 (9.1) | 0 (0) | |
| ALT in IU/L | 65.9 ± 30.5 | 98.9 ± 42.0 | 0.6541 |
| HBV-DNA (log) | 7.0 ± 1.60 | 6.87 ± 0.9 | 0.5263 |
| Number of amino acid residues mutations in S gene, n (%) | 34/41 (82.9) | 7/41 (17.1) | |
| 2001 | 2001 | 2003 | 2004 | |||||||||||||||
| No. | Sex | Age | ALT | HBVDNA | a-HBs | HBeAg | ALT | HBVDNA | a-HBs | HBeAg | ALT | HBVDNA | a-HBs | HBeAg | ALT | HBVDNA | a-HBs | HBeAg |
| 1 | M | 57 | 22 | 0 | - | - | 20 | 0 | - | - | 76 | 8.63 | - | + | 87 | 6.38 | + | + |
| 2 | M | 36 | 48 | 9.38 | - | + | 82 | 8.53 | - | + | 21 | 4.04 | - | + | 97 | 3.78 | + | + |
| 3 | F | 65 | 25 | 4.04 | + | - | 58 | 5.04 | + | - | 55 | 6.76 | + | - | 55 | 5.08 | + | - |
| 4 | M | 45 | 45 | 6.86 | + | + | 80 | 6.61 | + | + | 59 | 5.57 | - | + | 93 | 6.99 | + | + |
| 5 | M | 55 | 156 | 7.32 | + | + | 116 | 7.11 | + | + | 112 | 7.75 | + | + | 149 | 5.80 | + | + |
| 6 | F | 30 | 19 | 6.91 | + | + | 33 | 7.70 | + | + | 20 | 8.86 | - | + | 15 | 9.08 | + | + |
| 8 | M | 38 | 18 | 0 | + | - | 19 | 0 | + | - | 20 | 0 | - | - | 21 | 4.43 | + | - |
| 10 | M | 72 | 20 | 5.18 | + | + | 25 | 7.04 | + | + | 18 | 6.26 | - | + | 48 | 4.15 | + | + |
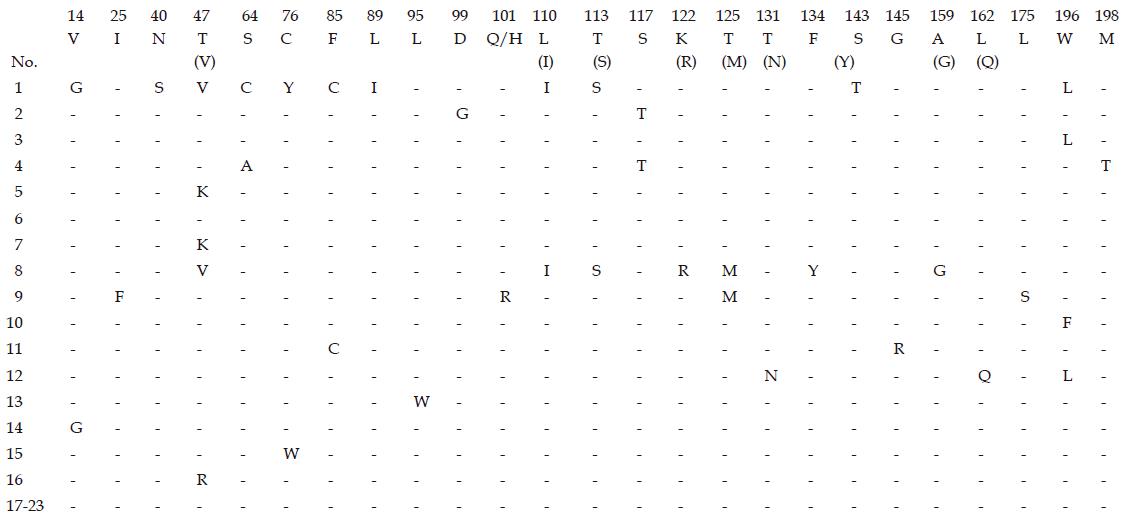
Nucleotide and deduced amino acid sequences of surface region and polymerase gene of HBV were performed in 23 patients. Comparison with the published HBV sequence showed that 21 (91.3%) out of 23 patients were infected with genotype C, 1 with genotype B and 1 with genotype D.15 (65.2%). Of the 23 patients who developed amino acid mutations in the surface gene protein, 10 were positive for anti-HBs and 5 were negative for anti-HBs. Mutations at the “α” determinant region were observed in 5 patients (5/15, 33.3%) (Figure 1). Forty-one mutations were found at 27 amino acid positions within the surface gene of HBV, and 34 mutations (82.9%, 34/41) were presented in the patients with coexisting HBsAg and anti-HBs. Six (14.6%) out of 41 mutations were located at the “α” determinant region, and 4 mutations were presented in the first loop (positions 124-137), the others were in the second loop (positions 139-147, S143T, G145R). Six mutations at amino acid residues 40 (N40S) and 47 (T47V, T47K, T47R) coincident with HLA class I-restricted (CTL) epitope[10] were observed in 5 patients, 11 mutations (26.8%) occurred in 6 patients within the major hydrophilic regions of upstream and downstream of the “α” determinant region (amino acid positions 99-169), 6 mutations at 3 amino acid positions (196, 198 and 199) associated with LMV-selected mutation were observed in 5 patients.
Because the S gene overlaps with the major calatytic domain of the polymerase gene, the mutations near the YMDD motif of the polymerase gene were studied. Eight mutations within amino acid residues 518-569 of the polymerase gene were observed at 4 positions (V173L, L180M, M204I/V, S223A) in 5 patients. Three patients who received long term LMV therapy and developed anti-HBs at the time of sequencing, had YMDD mutations (M204I/V) in polymerase gene and the S gene mutations at amino acid positions 196, 198 and 199 (Table 3).
| Mutantin patients | Position with HBsAg protein sequence change | Position with polymerase protein sequence change | ||||||
| (n) | 145 | 196 | 198 | 199 | 173 | 180 | 204 | 223 |
| Wild type | G | W | M | W | V | L | M | S |
| 1 | - | L | - | - | - | - | I | A |
| 3 | - | L | - | - | - | - | I | - |
| 10 | - | F | - | C | L | M | V | - |
| 11 | R | - | - | - | - | - | - | A |
| 13 | L | - | - | - | M | - | - | |
Five out of 15 (33.3%) patients who had amino acid mutations did not develop anti-HBs, while T131N, L162Q, W196L mutations in the S gene and L180M mutation in polymerase gene were simultaneously observed in only one of these patients.
HBV is the most common etiologic agent of chronic and often fetal liver diseases world wide. HBV variants present during natural infection or anti-virus therapy, and contribute to disease persistence. The S gene of HBV is crucial for binding and infectivity, and “α” determinant of the surface gene may form a target of humoral neutralizing antibody. Mutations in the region affect the binding of anti-HBs to corresponding HBsAg[5], produce escape from the neutralizing antibody, result in persistent infection and replication of HBV, even the relatively high titer anti-HBs develops.
In the present study, 15 patients had amino acid mutations in the surface gene of HBV, and 10 of them were positive for anti-HBs. Forty-one mutations were found in 23 patients, and 34 (82.9%) mutations were presented in the patients with coexisting HBsAg and anti-HBs. only 6 mutations within the “α” determinant region were observed in 5 patients (4 for anti-HBs positive). Passive or active immune therapy may develop the escape mutation[11,12], which has a point mutation from guanosine to adenosine at nucleotide 587 (condon 145, G145R). Mutation of G145R, however, was only seen in 1 patient in our study, and it may be the reason why no patient in our cohort received active or passive hepatitis B immunization. The data suggest that mutation within the “α” determinant region may play an important role in the presence of anti-HBs. A resent study showed that mutation in the “α” determinant region contributes to the most therapy failure, but there are still some therapy failures associated with mutations in the major hydrophilic region of the surface gene located at downstream or upstream of the “α” determinant (positions 99-169)[13]. In the present study, 26.8% mutations were observed in the above mention region, suggesting that these mutations also change the antigenicity of HBsAg and contribute to the development of anti-HBs. One patient who had amino acid mutation (T131N) in the “α” determinant region did not develop anti-HBs. It may be due to the relatively lower sensitive assays because it was reported that anti-Hbs complexed with HBsAg could be detected in nearly all patients with chronic hepatitis B when tested by a highly sensitive immunoassay[14].
Lamivudine-selected mutations in the S gene of HBV have been demonstrated by sequencing HBV isolated from the serum of patients treated with long-term LMV[15,16]. In addition, LMV-selected mutations within the HBsAg protein downstream of the “α” determinant (I195M, W196S and M198I) lead to a decrease in the antigenicity of the protein and binding to the anti-HBs antibodies, therefore poorly inhibiting their interaction with wild-type HBsAg[17]. In our study, the change of methionine to isoleucine (rtM204I) or valine (rtM204V) was found in the YMDD motif of the polymerase gene protein in three patients who received long-term LMV therapy. These patients also had W196L/W196F or W199C mutations within the surface gene protein of HBV simultaneously.
The surface gene of HBV also includes the putative HLA class I-restricted cytotoxic T lymphocyte (CTL) epitopes[2]. Because the putative CTL epitope mutations result in epitope inactivation and T cell receptor antagonism[18], mutant virus could evade cellular immunity and lead to persistent infection[19,20]. In the patients studied here, 6 mutations at amino acid residues 40 and 47 were observed in 1 and 5 patients, respectively, and 5 of them were positive for anti-HBs. These patients had relatively high HBV DNA level. Three of them had elevated ALT levels. This result is similar to the report from Taiwan, which revealed a high frequency of mutations at amino acid positions 40 and 47 of the surface gene in patients with chronic hepatitis B, suggesting that these mutations change CTL recognition and contribute to chronic infection in some patients[10].
In fact, patients positive for anti-HBs have more amino acid mutations, especially mutations in the crucial region of the surface gene associated significantly with the presence of anti-HBs. But presence of these mutations is not related to clinical features, ALT levels, HBV DNA levels and the presence of HBeAg, suggesting that these mutations may not alter the replicating competency of HBV although highly titer of anti-HBs develops. It is possible that these mutants may secrete into the serum via trans-complementation of intact protein in hepatocytes. The biochemical and virological follow-up parameters of 8 patients (No.1 to 6, No.8 and 10) showed that only 2 patients had persistent anti-HBs positive condition during the whole 4 year follow-up period, indicating that anti-HBs can be detected in chronic hepatitis B patients.
In conclusion, the presence of mutations in the “α” determinant of surface gene is not high in patients with coexisting anti-HBs and HBsAg. The mutations at the major hydrophilic region of the surface gene contribute to the development of anti-HBs in these patients and produce escape from the neutralizing antibody, and lead to persistent infection. Long-term LMV therapy could induce YMDD mutation in the polymerase gene and surface gene of HBV.
S- Editor Wang J L- Editor Wang XL E- Editor Bai SH
| 1. | Lok AS. Natural history and control of perinatally acquired hepatitis B virus infection. Dig Dis. 1992;10:46-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 2. | Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1189] [Cited by in RCA: 1202] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 3. | Chisari FV, Ferrari C. Hepatitis B virus immunopathology. Springer Semin Immunopathol. 1995;17:261-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 92] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 4. | Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi MG, Houghton M, Fiaccadori F, Ferrari C. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98:706-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 490] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 5. | Zheng X, Weinberger KM, Gehrke R, Isogawa M, Hilken G, Kemper T, Xu Y, Yang D, Jilg W, Roggendorf M. Mutant hepatitis B virus surface antigens (HBsAg) are immunogenic but may have a changed specificity. Virology. 2004;329:454-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Swenson PD, Escobar MR, Carithers RL Jr, Sobieski TJ 3rd. Failure of preexisting antibody against hepatitis B surface antigen to prevent subsequent hepatitis B infection. J Clin Microbiol. 1983;18:305-309. [PubMed] |
| 7. | Chen YC, Delbrook K, Dealwis C, Mimms L, Mushahwar IK, Mandecki W. Discontinuous epitopes of hepatitis B surface antigen derived from a filamentous phage peptide library. Proc Natl Acad Sci USA. 1996;93:1997-2001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 102] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25:97-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 146] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Ogura Y, Kurosaki M, Asahina Y, Enomoto N, Marumo F, Sato C. Prevalence and significance of naturally occurring mutations in the surface and polymerase genes of hepatitis B virus. J Infect Dis. 1999;180:1444-1451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 59] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Tai PC, Banik D, Lin GI, Pai S, Pai K, Lin MH, Yuoh G, Che S, Hsu SH, Chen TC. Novel and frequent mutations of hepatitis B virus coincide with a major histocompatibility complex class I-restricted T-cell epitope of the surface antigen. J Virol. 1997;71:4852-4856. [PubMed] |
| 11. | Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 772] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 12. | Carman WF, Trautwein C, van Deursen FJ, Colman K, Dornan E, McIntyre G, Waters J, Kliem V, Müller R, Thomas HC. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology. 1996;24:489-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 125] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Terrault NA, Zhou S, McCory RW, Pruett TL, Lake JR, Roberts JP, Ascher NL, Wright TL. Incidence and clinical consequences of surface and polymerase gene mutations in liver transplant recipients on hepatitis B immunoglobulin. Hepatology. 1998;28:555-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 113] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Maruyama T, McLachlan A, Iino S, Koike K, Kurokawa K, Milich DR. The serology of chronic hepatitis B infection revisited. J Clin Invest. 1993;91:2586-2595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 122] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Lok AS, Hussain M, Cursano C, Margotti M, Gramenzi A, Grazi GL, Jovine E, Benardi M, Andreone P. Evolution of hepatitis B virus polymerase gene mutations in hepatitis B e antigen-negative patients receiving lamivudine therapy. Hepatology. 2000;32:1145-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 150] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Ogata N, Fujii K, Takigawa S, Nomoto M, Ichida T, Asakura H. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in japanese patients with chronic hepatitis B. J Med Virol. 1999;59:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 17. | Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, Fyfe J, Sozzi T, Jackson DC. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293:305-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 170] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 18. | Bertoletti A, Sette A, Chisari FV, Penna A, Levrero M, De Carli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 407] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 19. | Aebischer T, Moskophidis D, Rohrer UH, Zinkernagel RM, Hengartner H. In vitro selection of lymphocytic choriomeningitis virus escape mutants by cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1991;88:11047-11051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Pircher H, Moskophidis D, Rohrer U, Bürki K, Hengartner H, Zinkernagel RM. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 461] [Article Influence: 13.2] [Reference Citation Analysis (0)] |