Published online Jun 7, 2006. doi: 10.3748/wjg.v12.i21.3373
Revised: September 28, 2005
Accepted: October 26, 2005
Published online: June 7, 2006
AIM: To clone Δ12 -fatty acid desaturase gene of Mortierella isabellina and to functionally characterize this gene in vitro and in vivo.
METHODS: Reverse transcriptional polymerase chain reaction (RT-PCR) was used to clone the open reading frame of Δ12-fatty acid desaturase gene (D12D) of Mortierella isabellina. Plasmids pEMICL12 and pYMICL12 were constructed with it. pEMICL12 was transformed into Escherichia coli (E.coli) strain BL21 using CaCl2 method for expression after induction with IPTG. pTMICL12 was transformed into Saccharomyces cerevisiae strain INVSc1 using lithium acetate method for expression under the induction of galactose. Northern blotting method was used to investigate the effect of temperature on the transcriptional level of this gene in S.cerevisiae strain INVSc1.
RESULTS: Recombinant plasmids pEMICL12 and pTMICL12 were successfully constructed and transformed into E.coli and S.cerevisiae separately with appropriate method. After induction with IPTG and galactose, it was found that expression of Δ12-fatty acid desaturase genes in E.coli and S. cerevisiae under appropriate conditions led to the production of active Δ12-fatty acid desaturase, which could convert 17.876% and 17.604% of oleic acid respectively to linoleic acid by GC-MS detection in vitro and in vivo.
CONCLUSION: Cloning and expression of M.isabellina D12D gene in E.coli and S.cerevisiae is successfully completed.
-
Citation: Li MC, Li H, Wei DS, Xing LJ. Cloning and molecular characterization of Δ12-fatty acid desaturase gene from
Mortierella isabellina . World J Gastroenterol 2006; 12(21): 3373-3379 - URL: https://www.wjgnet.com/1007-9327/full/v12/i21/3373.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i21.3373
As an economically important filamentous fungi, M isabellina which belongs to Mucor sp. can produce linolenic acid(GLA) that plays very important roles in regulating the hormone level and fatty acid metabolism in human. In addition, recent study indicated that GLA can alleviate the symptoms of many kinds of diseases such as neurodegeneration[1,2] , adipogenesis[3], cancer[4,5] , Protofibrils[6-8] , cardiovascular disease[9-11], diabetes[12] and psoriasis[13]. Being a kind of nutritionally and pharmaceutically important substance, GLA has become the hotspot both in academic and applied fields. M isabellina has been a widely used strain in industry to produce GLA by fermentation, in which GLA accounts for 8 percent of total fatty acids. Further efforts to enhance the percentage of the GLA by traditional mutation or by changing fermentation conditions such as medium and temperature have been made, but the extent is relatively small. Therefore, we attempted to reconstruct this strain with genetic engineering.
In most eukaryotic cells, GLA is biosynthesized by two desaturation processes, using stearic acid C18:1 as initial substrate[14-17] . Linoleic acid (LA) C18:2 is synthesized from C18:1 by the reaction catalyzed by Δ12-fatty acid desaturase and then LA can be converted to GLA by Δ6-fatty acid desaturase. LA can also be converted to α-linolenic acid (ALA) by ω3 fatty acid desaturase. LA and ALA are so-called essential fatty acids (EFAs)[18,19] to human bodies because of their inability to synthesize these compounds de novo. Linoleic acid is the precursor of polyunsaturated fatty acids (PUFAs) in both routes of n-6 and n-3[20]. Therefore, Δ12-fatty acid desaturase is the key enzyme for producing LA and is also the speed-limiting enzyme for routes of n-6 and n-3. Cloning and characterization of Δ12-fatty acid desaturase gene will help understand the biosynthetic pathway of GLA and further enhance the percentage of GLA in total fatty acids.
Our research interest focuses on PUFAs having 18 carbon atoms or more such as linoleic acid and linolenic acid in fungus. In this paper, we report the isolation of cDNA of D12D from M isabellina. The cDNA was expressed in a classical system, S.cerevisiae and E.coli.
Mortierella isabellina M 6-22 was grown at 28°C for 2 d in liquid medium containing 2% glucose, 1% bacto-yeast extract, 0.2% KH2PO4 and 0.1% MgSO4, with pH adjusted to 6.0. S. cerevisae strain INVScI was used as recipient strain in transformation experiments and was grown at 30°C in complex medium containing 1% bacto-yeast extract, 2% bacto-peptone and 2% glucose. E.coli strain DH5a was grown at 37°C in Luria–Bertani medium (LB) supplemented with 100 mg/L of ampicillin. E.coli strain BL21 and the expression vector pET21a were purchased from Novegon(Novegen Inc., Madison, WI).
Restriction endonucleases, and the other DNA-modifying enzymes were obtained from TaKaRa Bio. Dalian, China Co. Ltd. Other chemicals were purchased from Sangon Bio. Shanghai, China Co. Ltd.
Mycelia were harvested by filtration and washed with phosphate buffer. The extraction of total RNA was done according to the guanidinium thiocyanate method as described by Chomczynski and Sacchi[21] and extracts were stored at -70°C for future use.
A pair of primers was designed according to the reported sequence of Δ12-fatty acid desaturase gene from Mortierella alpine[22] to clone the open reading frame (ORF) of the M isabellina D12D gene, P1: 5’-TAC CTC GAA TCC ATG GCA CCT CCC AAC ACT ATC GAT GCC-3’; P2: 5’-AAC CGT CTC GAG TTA CTT CTT GAA AAA GAC CAC GTC TCC-3’, which corresponded to the nucleotide sequences of start and stop codons (in boldface) of the Δ12-fatty acid desaturase gene, respectively. The 5’-end of both primers contained an EcoRIand an XhoIsite(underlined and in boldface), respectively that were underlined to facilitate subsequent manipulation. PCR was performed in a Biometra T-gradient thermal cycler using the following program: at 94°C for 1 min, 53°C for 1 min, 72°C for 2 min for 30 cycles, followed by extension for 10 min at 72°C. PCR fragments were subcloned into pGEM-T vector (Promega, Madison, WI) to produce recombinant plasmid pTMICL12, which was then transformed into E.coli DH5a. Subsequently, nucleotide sequences were determined (TaKaRa Bio, Dalian, China). Analysis of the sequences was done with DNAMAN software (version 4.0, Lynnon BioSoft, Quebec).
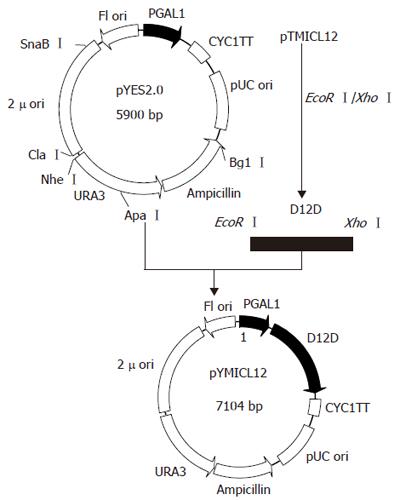
The ORF of Δ12-fatty acid desaturase from M isabellina was digested from pTMICL12 with EcoRIand XhoIand subcloned respectively into S.cerevisiae expression vector pYES2.0 and E.coli expression vector pET21a. Positive transformants were screened on the LB plate with ampicillin resistance. The resultant positive plasmid was validated by PCR and restriction enzyme digestion. The construction sketch of pYMICL12 is shown in Figure 1. The resultant plasmids, pYMICL12 and pEMICL12 were stored and propagated in E.coli strain DH5a.
The recombinant plasmid pYMICL12 was transformed into S.cerevisiae INVSc1 using the lithium acetate method [23]. Positive transformants were selected by plating on complex synthetic minimal medium agar lacking uracil (SC-Ura) and grown at 30°C for 2-3 d. The putative Δ12-desaturase gene was heterologously expressed in yeast, which was induced under transcriptional control of the GAL1 promoter. Yeast cultures were grown to logarithmic phase at 30°C in synthetic minimal medium containing different concentrations of galactose(1%, 2%, 3%, 4% respectively), and 0.67% yeast nitrogen. Subsequently, cells were harvested by centrifugation followed by washing in sterile water for three times. The cells were dried and ground into fine powders for determination of fatty acid composition by gas chromatography and for gas chromatography–mass spectrometry (GC-MS) analysis.
Transformant containing plasmid pETMICL12 was cultured in M9 medium at 37°C overnight. Five hundred milliliter M9 medium was inoculated into 2.8 L Fernbach flask with aliquot from the overnight culture until the light density reached 0.5. IPTG was added until the final concentration reached 0.4 mmol/L. After 4 hours’ induction, cells were pelleted by centrifugation at 5000 r/min for 10 min at 4°C and stored at -70°C for future use.
Cells defrosted at room temperature were suspended in 30 mL buffer A (containing 10 mmol/L MgCl2 , 50 mmol/L MOPS-NaOH, 1 mmol/L EGTA, 10 μg/mL DNase) and then were treated with supersonic 450 Hz for 40 cycles, 4 s per cycle. Intact cells and cell debris were pelleted by centrifugation at 5000 r/min for 10 min at 4°C. Supernatants containing cell membrane were pelleted by super-centrifugation at 150 000 r/min for 30 min at 4°C and then were re-suspended in 200 μL buffer A.
Ten percent of the gel was used to separate the protein bands, and stained by Coomassie Blue R-250[24].
Sodium salt was added into the membrane suspension in buffer A to reach the final concentration of 150 μmol/L and then the mixture was incubated at 25°C in water bath for 30 min after oleic acid was added. Soon after that, the mixture was extracted with chloroform methanol and water (volume ratio 2:1:2). The extraction was methyl-esterified with 5% HCl/methanol at 85°C for 150 min. Fatty acid methyl ester was solubilized with hexane[25,26].
Total fatty acid was extracted from the cells by treating 100 mg yeast powder with 5 mL 5% KOH in methanol for saponification at 70°C for 5 h. pH of the product was adjusted to 2.0 with HCl (6 mol/L) before the fatty acid was methyl-esterified with 4 mL 14% boron trifluoride in methanol at 70°C for 1.5 h. Then, fatty acid methyl esters (FAME) were solubilized with hexane after addition of saturated sodium chloride solution. FAMEs were analyzed by gas chromatography (GC; GC-9A, Shimadzu, Kyoto, Japan) and identified by the comparison of their peaks with that of standards (Sigma). Qualitative analysis of FAME was performed by GCMS using an HP G1800A GCD system (Hewlett–Packard, Palo Alto, CA, USA). Both analyses were carried out with the same polar capillary column (HP, 5.30 mU, 0.25 mm in internal diameter). The mass spectrum of a new peak was compared with that of the standard for identification of fatty acid.
Northern blotting was used to investigate the effect of temperature on the transcriptional level of MID12. Total RNA of transgenic yeast after induction at different temperatures was extracted by UNIQ-10 column (Sangon Corp.) and digoxingenin (Roche Diagnostics Corp.) was used to label the DNA probe (PCR of MID12)
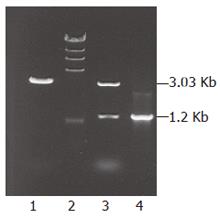
PCR product about 1.2 kb long was gel-purified by electrophoresis and cloned into pGEM-T vector. Positive clones were selected on LB plate by color reaction. PCR and enzyme restriction were used to analyze the resultant transformant named pTMICL12. After digestion with SacIIand PstI, a 3.03 kb fragment and a 1.2 kb target gene were obtained as shown in Figure 2. Subsequently, nucleotide sequences were determined (TaKaRa Bio, Dalian, China)
Nucleic acid sequence of Δ12 -fatty acid desaturase in M. isabellina has been deposited in Genbank database and assigned the accession No. AF417245. Comparison of the DNA sequence with other Δ12-fatty acid desaturase gene showed that it has the highest identity with Δ12-fatty acid desaturase gene from M. alpina (about 99.92%) and higher identity with other D12D from fungi[27-30] (more than 40%). Therefore, this sequence encoded a putative Δ12-fatty acid desaturase gene.
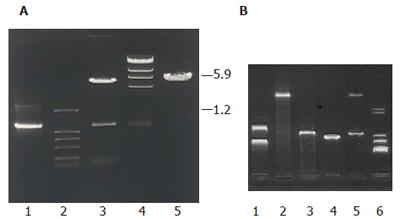
In order to investigate the function of this sequence, target gene was subcloned into YES2.0 and pET21a to construct two recombinant plasmids pYMICL12 and pEMICL12 which were transformed into E.coli DH5a for propagation. After digested by EcoRI and XhoI, both of the plasmids released a 1.2 kb fragment (Figure 3), indicating that the recombinant plasmids were successfully reconstructed.
After inducement, the total fatty acid in transgenic yeast harboring plasmid pYMICL12 was extracted and analyzed by GC. It was found that under different conditions, the expressed desaturase showed different activities.
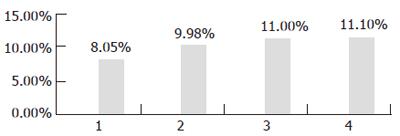
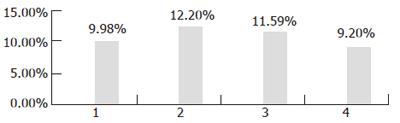
First, the transgenic yeast was induced at 30°C for 36 h, with different concentrations of galactose: 1%, 2%, 3%, 4%. The result of GC showed that percentage of LA in transgenic yeast was the highest when it was induced with 3% galactose (Figure 4).
Then, different duration times were used to induce the transgenic yeast by 3% galactose at 30°C for 12, 24, 36, and 48 h respectively. The result of GC showed that the optimal induction time was 24 h as shown in Figure 5.
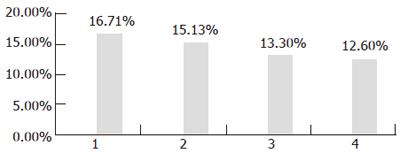
Thirdly, the transgenic yeasts were induced by 3% galactose for 24 h at different temperatures of 15°C, 20°C, 25°C, and 30°C respectively. It was found that the lower the temperature, the higher the activity of the expressed enzyme. The highest percentage of LA reached 16.706% of total fatty acid in induced yeast harboring plasmid pYMICL12 when the temperature was 15°C (Figure 6).
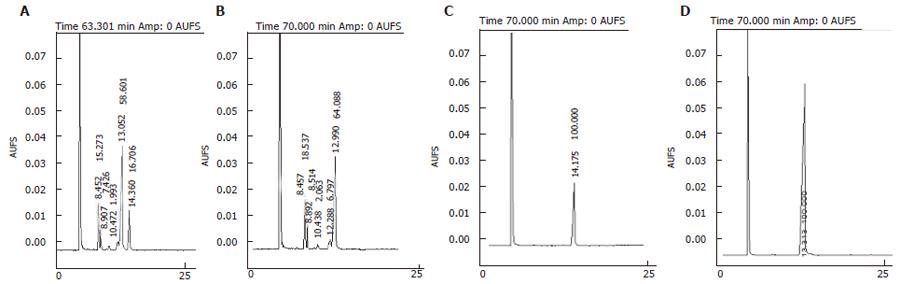
Therefore, the optimal condition for expressing D12D from M isabellina was: 3% galactose induction for 24 h at 15°C. GC analysis of the fatty acid composition of the transgenic yeast containing plasmid pYMICL12 indicated that a novel peak that had the same retention time with that of LA standard appeared. That specific peak was absent from the control transformant (Figure 7).
Northern blot was performed to check if there was an increase of mRNA after induction by low temperature which resulted in the higher activity of Δ12-fatty acid desaturase in transgenic yeast, INVSc1(pYMICL12).
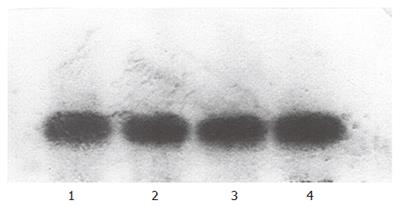
After induction at different temperatures, total RNA was extracted. In the formaldehyde-denaturalization electrophoresis, 30 mg RNA was used for each sample. The result of the Northern blot is shown in Figure 8.
Any visible change of the level of RNA could not be found on the photo of Northern blot. Analysis by VDS-scan also showed no obvious difference between the samples, implying there were no changes of the mRNA level of D12D in the transgenic yeast at different temperature. The level of transcription was not the key cause for the change of LA level in INVSc1 (pYMICL12) under cold condition. The increasing activity of Δ12-fatty acid desaturase might come from the protection of the enzyme activity by low temperature, which even prolonged the half life of Δ12-fatty acid desaturase mRNA.
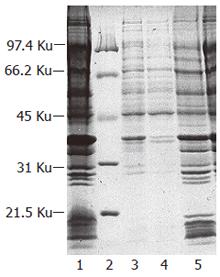
Transformants harboring plasmid pEMICL12 were cultured in M9 medium and induced by IPTG to express Δ12-fatty acid desaturase from M isabellina. Cell membranes were pelleted by super-centrifugation. Analysis with SDS-PAGE showed that a specific protein band that had the molecular weight of approximately 43 kd existed on the gel, whereas cell membrane from control transformant harboring the plasmid pET21a did not express the specific protein. In addition, the molecular weight of the specific protein band was nearly identical to other reported Δ12-fatty acid desaturase. These results suggested that this specific band putatively was Δ12-fatty acid desaturase (Figure 9).
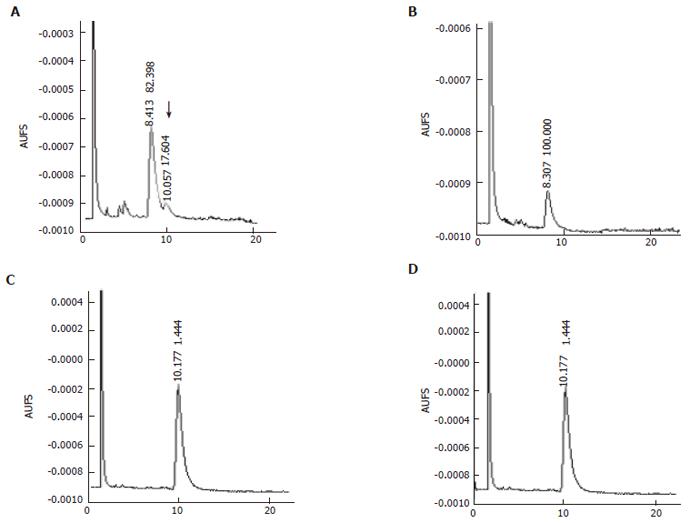
In order to test whether this protein band could convert OA to LA, in vitro assay with membrane protein was done. E coli can not produce OA that is the most commonly used substrate of Δ12-fatty acid desaturase. Therefore, OA must be added into the reaction mixture to be converted to LA by Δ12-fatty acid desaturase. GC analysis of the mixture after reaction indicated that transformant containing plasmid pETMICL12 exhibited Δ12-fatty acid desaturase activity and that the membrane protein could convert 17.604% of the OA to LA(indicated by arrow), whereas cell membrane protein from transformant containing empty plasmid pET21a did exhibit Δ12-fatty acid desaturase activity. Also, the retention time of the specific peak in Figure 10 was identical to standard LA from Sigma. These results indicated that the specific band had Δ12-fatty acid desaturase activity.
M isabellina strain M6-22 is a mutant strain in which total fatty acids account for 60 percent of the dry weight of the mycelium. Also, it synthesizes PUFAs only up to 18-carbon with c isomer of linolenic acid (GLA), which make the subsequent purification relatively easy. Thus, M isabellina strain M6-22 is a promising producer of GLA and also a potential strain to be used to produce other nutritionally and pharmaceutically important PUFAs. However, increasing the percentage of GLA meets great challenges, which made us to turn to other methods including genetic engineering. As we all know, cloning the genes encoding corresponding enzymes is the first thing we should do. In our previous work, we have successfully cloned and characterized Δ6-fatty acid desaturase gene from M isabellina[31]. In order to further elucidate the biosynthetic pathway of polyunsaturated fatty acid, we decided to clone the enzyme genes that are ahead of the Δ6-fatty acid desaturase gene in reaction chains.
As M isabellina is very near to M alpine in phylogeny, we adopted the strategy of direct cloning the gene by designing the primer identical to the reported M alpineΔ12-fatty acid desaturase gene. Using this method, a fragment of proximately 1.2 kb was obtained. Sequence comparison indicated it has the highest identity with M alpineΔ12-fatty acid desaturase gene. In order to functionally identify this nucleic acid sequence, two expression systems were applied: yeast and E.coli. In both systems, the heterologously expressed proteins exhibited Δ12-fatty acid desaturase activity, indicating that this sequence encoded a functional desaturase. The high identity between the two sequences suggests that M alpine and M isabellina belong to the same species. But there indeed exist great differences between the two species. M alpine can produce many kinds of PUFAs, but M isabellina can only synthesize PUFAs up to 18-carbon. This indicates that other factors may contribute to the difference.
Protein expression in yeast is influenced by many factors such as temperature, induction time. In this work, different temperatures were used. In a certain extent (15-30°C), the higher the temperature, the lower the desaturase activity. At even lower temperature, the yeast INVSc1 (pYMICL12) could achieve more LA. However, a temperature lower than 15°C would hinder the growth of the mycelium, which reduced the total activity of Δ12 -fatty acid desaturase.
Although desaturase activity increased as the temperature decreased, northern blotting analysis indicated that the increasing of desaturase activity was not due to the corresponding increase of the mRNA level. In our study, no significant difference existed among the mRNA level at different temperatures. Other researchers also have reported the same result[28]. This indicates that under certain conditions, mRNA level can not be influenced by temperature. The most important factor that can change it may be the promoter activity. Does the increased activity of Δ12-fatty acid desaturase, however, come from the influence of coldness on mRNA level or protein level Or does longer half life of Δ12-fatty acid desaturase even directly influence activity of protein There are different opinions about this in academia.
In conclusion, this report about cloning and characterization of the M isabellinaΔ12-fatty acid desaturase gene will be helpful for us to utilize the industrially used strains.
S- Editor Wang J L-Editor Zhu LH E- Editor Zhang Y
| 1. | Lansbury PT Jr. Back to the future: the 'old-fashioned' way to new medications for neurodegeneration. Nat Med. 2004;10 Suppl:S51-S57. [PubMed] [DOI] [Full Text] |
| 2. | Rochet JC, Outeiro TF, Conway KA, Ding TT, Volles MJ, Lashuel HA, Bieganski RM, Lindquist SL, Lansbury PT. Interactions among alpha-synuclein, dopamine, and biomembranes: some clues for understanding neurodegeneration in Parkinson's disease. J Mol Neurosci. 2004;23:23-34. [PubMed] [DOI] [Full Text] |
| 3. | Ray SS, Lansbury PT Jr. A possible therapeutic target for Lou Gehrig's disease. Proc Natl Acad Sci U S A. 2004;101:5701-5702. [PubMed] [DOI] [Full Text] |
| 4. | Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT Jr. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837-846. [PubMed] [DOI] [Full Text] |
| 5. | Lane J, Mansel RE, Jiang WG. Expression of human delta-6-desaturase is associated with aggressiveness of human breast cancer. Int J Mol Med. 2003;12:253-257. [PubMed] |
| 6. | Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267-298. [PubMed] [DOI] [Full Text] |
| 7. | Park JY, Lansbury PT Jr. Beta-synuclein inhibits formation of alpha-synuclein protofibrils: a possible therapeutic strategy against Parkinson's disease. Biochemistry. 2003;42:3696-3700. [PubMed] [DOI] [Full Text] |
| 8. | Anguiano M, Nowak RJ, Lansbury PT Jr. Protofibrillar islet amyloid polypeptide permeabilizes synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry. 2002;41:11338-11343. [PubMed] [DOI] [Full Text] |
| 9. | Lauritzen L, Hansen HS, Jørgensen MH, Michaelsen KF. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog Lipid Res. 2001;40:1-94. [PubMed] [DOI] [Full Text] |
| 10. | Kris-Etherton P, Daniels SR, Eckel RH, Engler M, Howard BV, Krauss RM, Lichtenstein AH, Sacks F, St Jeor S, Stampfer M. Summary of the scientific conference on dietary fatty acids and cardiovascular health: conference summary from the nutrition committee of the American Heart Association. Circulation. 2001;103:1034-1039. [PubMed] [DOI] [Full Text] |
| 11. | Curtis CL, Hughes CE, Flannery CR, Little CB, Harwood JL, Caterson B. n-3 fatty acids specifically modulate catabolic factors involved in articular cartilage degradation. J Biol Chem. 2000;275:721-724. [PubMed] [DOI] [Full Text] |
| 12. | Shaw JT, Purdie DM, Neil HA, Levy JC, Turner RC. The relative risks of hyperglycaemia, obesity and dyslipidaemia in the relatives of patients with Type II diabetes mellitus. Diabetologia. 1999;42:24-27. [PubMed] [DOI] [Full Text] |
| 13. | Mayser P, Grimm H, Grimminger F. n-3 fatty acids in psoriasis. Br J Nutr. 2002;87 Suppl 1:S77-S82. [PubMed] [DOI] [Full Text] |
| 14. | Xian M, Nie J, Meng Q, Liu J, Zhou C, Kang Y, Zhen K. Production of gamma-linolenic acid by disrupted mycelia of Mortierella isabellina. Lett Appl Microbiol. 2003;36:182-185. [PubMed] [DOI] [Full Text] |
| 15. | Xian M, Kang Y, Yan J, Liu J, Bi Y, Zhen K. Production of linolenic acid by Mortierella isabellina grown on octadecanol. Curr Microbiol. 2002;44:141-144. [PubMed] [DOI] [Full Text] |
| 16. | Murphy DJ. Biogenesis, function, and biotechnology of plant storage lipids. Prog Lipid Res. 1994;33:71-85. [PubMed] [DOI] [Full Text] |
| 17. | Kinney AJ, Cahoon EB, Hitz WD. Manipulating desaturase activities in transgenic crop plants. Biochem Soc Trans. 2002;30:1099-1103. [PubMed] [DOI] [Full Text] |
| 18. | Gill I, Valivety R. Polyunsaturated fatty acids, Part 1: Occurrence, biological activities and applications. Trends Biotechnol. 1997;15:401-409. [PubMed] [DOI] [Full Text] |
| 19. | Innis SM. Essential fatty acids in growth and development. Prog Lipid Res. 1991;30:39-103. [PubMed] [DOI] [Full Text] |
| 20. | Alonso DL, Maroto FG. Plants as 'chemical factories' for the production of polyunsaturated fatty acids. Biotechnol Adv. 2000;18:481-497. [PubMed] [DOI] [Full Text] |
| 21. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [PubMed] [DOI] [Full Text] |
| 22. | Sakuradani E, Kobayashi M, Ashikari T, Shimizu S. Identification of Delta12-fatty acid desaturase from arachidonic acid-producing mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur J Biochem. 1999;261:812-820. [PubMed] [DOI] [Full Text] |
| 23. | Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genomics: A Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press 1998; . |
| 24. | Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd ed. New York: Cold spring Harbor Laboratory Press 1993; 463-468. |
| 25. | Sakuradani E, Kobayashi M, Shimizu S. Delta 9-fatty acid desaturase from arachidonic acid-producing fungus. Unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur J Biochem. 1999;260:208-216. [PubMed] [DOI] [Full Text] |
| 26. | Panpoom S, Los DA, Murata N. Biochemical characterization of a delta12 acyl-lipid desaturase after overexpression of the enzyme in Escherichia coli. Biochim Biophys Acta. 1998;1390:323-332. [PubMed] [DOI] [Full Text] |
| 27. | Passorn S, Laoteng K, Rachadawong S, Tanticharoen M, Cheevadhanarak S. Heterologous expression of Mucor rouxii delta(12)-desaturase gene in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1999;263:47-51. [PubMed] [DOI] [Full Text] |
| 28. | Watanabe K, Oura T, Sakai H, Kajiwara S. Yeast Delta 12 fatty acid desaturase: gene cloning, expression, and function. Biosci Biotechnol Biochem. 2004;68:721-727. [PubMed] [DOI] [Full Text] |
| 29. | Pain A, Woodward J, Quail MA, Anderson MJ, Clark R, Collins M, Fosker N, Fraser A, Harris D, Larke N. Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fungal Genet Biol. 2004;41:443-453. [PubMed] [DOI] [Full Text] |
| 30. | Wei D, Li M, Zhang X, Ren Y, Xing L. Identification and characterization of a novel delta12-fatty acid desaturase gene from Rhizopus arrhizus. FEBS Lett. 2004;573:45-50. [PubMed] [DOI] [Full Text] |
| 31. | Li MC, Liu L, Hu GW, Xing LJ. [Expression of Mortierella isabellina delta6-fatty acid desaturase gene in gamma-linolenic acid production in transgenic tobacco]. Shengwu Gongcheng Xuebao. 2003;19:178-184. [PubMed] |