INTRODUCTION
As one of the signal transducers and activators of transcription (STAT) family members, STAT3 has been correlated with positive regulation of cell growth and is highly activated in cancer cells[1,2]. Activation of STAT3 signaling regulates the expression of numerous genes involved in growth control and survival. Many studies have shown that numerous genes encoding Bcl-xL, Mcl-1, cyclins D1/D2, and c-Myc proteins are downstream targets of STAT3[3-6]. Recent studies have indicated that constitutive STAT3 signaling induces VEGF expression and tumor angiogenesis[7-11]. The expression of VEGF antigen in gastric cancer cells can serve as a pertinent predictive factor for hematogenous invasion or metastasis, and the importance of it has been proven and widely studied[12-14]. In addition, the resistance to 5-fluorouracil (5-FU) is a main obstacle in gastric cancer chemotherapy. However, the correlation between STAT3 and VEGF has not been studied in drug resistant cell line. In this study, we aimed to investigate the different activation of STAT3 signaling in two human stomach adenocarcinoma cell lines, 5-fluorouracil resistant cell line and its parental cell line, and evaluate its relationship with the expression of VEGF.
MATERIALS AND METHODS
Reagents
Nuclear and cytoplasmic extraction reagents and bovine serum albumin (BCA) protein assay kit were purchased from Pierce (Rockford, IL, USA). Easytides adenosine 5’-triphosphate [γ-32P] was purchased from PerkinElmer (Boston, MA, USA). MicroSpinTM G-25 column and hybond-C membrane were purchased from Amersham Biosciences (Piscataway, NJ USA). TRIzol reagent and MagicMarker Western standard were purchased from Invitrogen (Carlsbad, California. USA). AMV reverse transcription system (A3500), Taq DNA polymerase, dNTP, PCR marker and EMSA kit were purchased from Promega (Madison, WI, USA). Monoclonal anti-β-actin was purchased from Sigma. Polyclonal rabbit anti-human phospho-Tyr705-STAT3 was obtained from Cell Signaling (Beverly, MA, USA). Goat anti-rabbit IgG-AP was purchased from Santa Cruz (California, USA). Monoclonal mouse anti-human VEGF antibody was obtained from Fujian Maixin Co (Fujian, China). DAKO Envision system HRP (DAB) was purchased from DAKO (Produktionsvej, Glostrup, Denmark).
Cell lines and culture
Two human gastric adenocarcinoma cell lines, 5-fluorouracil resistant cell line SGC7901/R and its parental cell line SGC7901 were obtained from Shanghai Institute of Materia Medica, Chinese Academy of Sciences. All of the cells were grown in RPMI 1640 supplemented with 100 mL/L fetal bovine serum in a humidified atmosphere containing 50 mL/L CO2 at 37 °C. The final inducing dosage of 5-fluorouracil was 1 g/L.
Western blot analysis
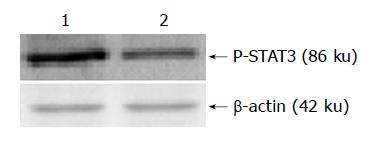
Both the extraction of nuclear protein of two cell lines and the contents of nuclear protein were determined by the kits according to the manufacturer’s instruction. A total of 100 μg of nuclear extracts was loaded onto SDS-polyacrylamide gel and blotted onto hybond-C membranes by electrophoresis. Equal protein sample loading was monitored by hybridizing the same membrane filter with an anti-β-actin antibody. MagicMarker was used for molecular weight determinations. The membranes were rinsed in TBS and blocked for 1 h at room temperature with 50 g/L fat free milk powder in TBS. The membranes were then incubated with 1:400 dilution of polyclonal antibodies against phospho-STAT3 at room temperature for 2 h. The proteins were detected by incubating the strips in alkaline phosphatase-conjugated anti-rabbit IgG antibody for 1 h at room temperature. The Quantity one software was used to analyse the scanned protein bands.
Electrophoretic mobility shift assay (EMSA) for STAT3-DNA binding activity
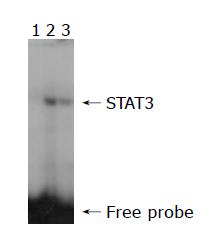
The STAT3-DNA binding activity was assessed by EMSA using the nuclear extract from the two cell lines. The sense strand that binds activated STAT3 protein was 5’ TCGACATTTCCCGTAAATC 3’ (synthesized by Shanghai Shenggong Co). Double-stranded oligonucleotide was end-labeled with [γ-32P] ATP using a T4 polynucleotide kinase according to the manufacturer’s instruction. The final concentration of probe was 1.75 pmol/L. The labeled probes were then purified by G-25 spin columns. One microliter of 32P-labeled STAT3 oligonucleotide was added to each reaction. For STAT3 specific test, a 150-fold unlabeled STAT3 probe was applied as a competitor. The final volume of reaction was 20 μL, including 10 μg of nuclear extract and 5×binding buffer. The reactions were placed on ice for 30 min. The 45 g/L nondenaturing acrylamide gel was pre-run in 1×TBE buffer at 25 mA for 60 min. After loading of the samples, the gel was run at room temperature in 1×TBE buffer at 25 mA for 90 min. The gel was dried on a gel dryer, then exposed to x-ray film overnight at -80 °C with intensifying screen. The protein-DNA complex was detected by autoradiography. The Quantity one software was used to analyse the scanned EMSA gel bands.
Semi-quantitative RT-PCR
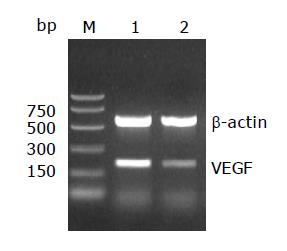
Total RNA was extracted from the two cell lines with a single step method using the TRIzol reagent according to the manufacturer’s protocol. Complementary DNA (cDNA) synthesis was performed using 2 μg of total RNA by reverse transcription system (A3500) according to the manufacturer’s instructions. Polymerase chain reaction was then conducted for 26, 28, 30 and 35 cycles in a thermal controller (Programmable Thermal Controller; MJ Research, Inc.) and the optimal cycle number for quantification (30 cycles) was determined. The primer annealing for STAT3 was carried out at 57 °C for 0.5 min. The sequences of PCR primers were as follows: for VEGF, 5’ TGCATTCACATTTGTTGTGC 3’(sense), and 5’ AGACCCTGGTGGACATCTTC 3’ (antisense, a 200 bp product); for β-actin, 5’ AAGGATTCCTATGTGGGC 3’ (sense), and 5’ CATCTCTTGCTCGAAGTC 3’ (antisense, a 532 bp product). β-actin mRNA levels were used as internal controls. Then 8 μL of each PCR product was electrophoresed on 20 g/L agarose gels, and the intensities of the specific bands were analyzed by the software of Quantity one.
Immunocytochemistry
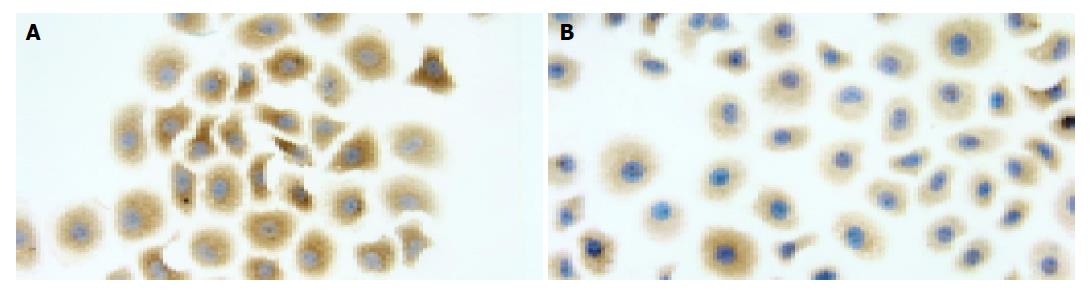
DAKO Envision system HRP (DAB) was used for immunocytochemical staining of VEGF. Two kinds of cells were fixed in 40 g/L paraformaldehyde. Endogenous peroxidase and nonspecific background staining were blocked by incubating slides with 30 mL/L hydrogen peroxide for 10 min at room temperature. After washed with PBS for 5 min, slides were blocked with normal serum for 20 min, followed by incubation with the primary antibody for VEGF (a mouse anti-human monoclonal antibody diluted at 1:50), overnight at 4 °C. As negative controls, PBS was used to replace primary antibody. After rinsed with PBS for 5 min, slides were incubated with the secondary antibody for 30 min and washed again. Chromogen was developed with 3, 3’-diaminobenzidine (DAB). All of the slides were lightly counterstained with hematoxylin for 10 s before dehydration and mounting. The slides were examined under a microscope (BX-50, Olympus Company). The software of Spot version 3.5.2 was used to analyze the images.
DISCUSSION
The STAT factors function as downstream effectors of cytokine and growth factor receptor signaling. STAT signaling is critical for normal cellular processes, such as embryonic development, organ genesis and function, innate and adaptive immune function, regulation of cell differentiation, growth, and apoptosis. Studies have demonstrated constitutive activation of STAT, in particular STAT1, STAT3, and STAT5, in a large number of diverse human tumor cell lines. The function of Stat1 has been associated with growth suppression rather than malignant transformation and thus can be considered a potential tumor suppressor. However, STAT3 and STAT5, have been demonstrated to directly contribute to oncogenesis by stimulating cell proliferation and preventing apoptosis. A variety of cellular mechanisms of multi-drug resistance (MDR) have been identified in human drug-resistant cell lines. These include P-glycoprotein (Pgp), multi-drug resistance-associated protein (MRP), lung resistance-related protein (LRP), glutathiones-transferase (GST-pi), DNA topo-isomerase II alpha (Topo II alpha) and apoptosis, etc. Catlett-Falcone et al[16 ] found that constitutive activation of STAT3 signaling conferred resistance to apoptosis in human U266 myeloma cells. But inhibition of STAT3 signaling led to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. These studies suggested that tumor cells lacking STAT activation were typically more tolerant to the pharmacological doses used in these experiments[17].
A main chemotherapeutic drug for patients with progressive gastric cancer 5-fluorouracil is one of the pyrimidine antagonists. A research on 5-FU resistant human gastric cancer cell line showed that SGC7901/R was an acquired resistant cell line[18]. Besides highly resistant to 5-FU, the drug-resistant cell line was cross-resistant to several drugs. SGC7901/R cell line was 12.77, 271 and 14.04-fold more resistant to cytotoxic action of DNR, CDDP and MMC, respectively. So the 5-FU resistant cell line has the characteristics of multi-drug resistance (MDR). The drug resistance mechanisms of 5-FU resistant cell line were related not only to the alteration of thymidylate synthase (TS) and thymidine kinase (TK), which play key role in the pathway of pyrimidine synthesis, but also to the P-glycoprotein (P-gp) coded by mdr-1 genes.
The present study originally investigated the relationship between STAT3 and MDR in the drug-resistance cell line. Western blot was firstly used to detect the expression of phospho-STAT3 protein. The result demonstrated that the expression of phospho-STAT3 protein was lower in the drug-resistant cell line SGC7901/R as compared with its parental cell line SGC7901. The EMSA assay provides a simple and rapid method for detecting DNA-binding protein. This method has been used widely in the study of sequence-specific DNA-binding proteins, such as transcription factors. The result of EMSA in our study showed that STAT3 DNA-binding activities were different between the two cell lines. The STAT3 DNA-binding activity was decreased in the drug-resistant cell line. These evidence suggested that drug-resistant cells which had lower level of STAT activation was more tolerant to the drugs of chemotherapy.
Numerous studies on STAT signal pathway have shown that signals usually generated at cell surface receptors. Upon binding of cytokines to cognate receptors on the surface of cells, receptors dimerize and thereby activate receptor-associated tyrosine kinases, such as JAKs. Receptor-bound STAT subsequently become tyrosine phosphorylated either by receptor intrinsic or associated tyrosine kinase activities. Once phosphorylated, STAT dimerizes via reciprocal phosphotyrosine-SH2 domain interactions. Within minutes, the dimers translocate to the nucleus, where they interact with other transcriptional modulators bound to specific promoter sequences and induce gene expression. Wei et al[19] found that more than 80% of the human pancreatic cancer cell lines exhibited constitutively activated STAT3, with STAT3 activation correlated with the VEGF expression level. Immunohistochemical studies have indicated that VEGF over-expression coincides with elevated STAT3 activation in human pancreatic cancer specimens. Blockade of activated STAT3 via ectopic expression of dominant-negative STAT3 significantly suppressed VEGF expression, angiogenesis, tumor growth, and metastasis in vivo. Furthermore, constitutively activated STAT3 directly activated the VEGF promoter, whereas dominant-negative STAT3 inhibited the VEGF promoter. These results indicated that STAT3 directly regulated VEGF expression and hence angiogenesis, growth, and metastasis of human pancreatic cancer. The present study demonstrated that compared with the parental cell line SGC7901, both the constitutive activation of STAT3 and the expressive intensity of phospho-STAT3 protein were lower in drug resistant cell line. The expression levels of VEGF mRNA and its encoded protein were also decreased in drug-resistant cell line. These results suggested that over-expression of VEGF might be correlated with elevated STAT3 activation in parental cell line and the lower VEGF expression is correlated with decreased STAT3 activation in resistant cell line.
A number of modulators of STAT signaling pathways have been described[20,21], and the suppressor of cytokine signaling has received a great deal of focus. The suppressors of cytokine signaling (SOCS) family consist of eight proteins, CIS and SOCS1-SOCS7, which were discovered to suppress STAT signaling by binding to and inhibiting JAKs[22-24]. Some of these proteins were transcriptionally regulated by STAT themselves, suggesting that STAT could negatively regulate their own phosphorylation state. SOCS1 could play a key role in the negative regulation of interferon-gamma signaling and in T cell differentiation[25]. It has been demonstrated to block Tel-JAK2-mediated transformation of hematopoietic cells[26]. Recently, a deletion on chromosome 16p that contains SOCS1 was found in 48% of primary hepatocellular carcinomas, raising the possibility that inactivation of this gene might participate in hepatocarcinogenesis[27]. SOCS2 (-/-) mice were 30-40% larger than wild-type mice, demonstrating that SOCS2 might be a critical regulator of postnatal growth[24]. SOCS-3, which is known to inhibit STAT3 and STAT5 activation, has been shown to be critical in the negative regulation of fetal liver hematopoiesis[28]. The biological role of other SOCS proteins remains to be determined. The protein inhibitors of activated STAT (PIAS) represent another group of proteins that normally serve to decrease DNA activation by blocking STAT DNA binding activity[29]. Reports have demonstrated that over-expression of PIAS3 suppresses gene transcription mediated by STAT3[30]. The present study demonstrated that both the STAT3-DNA binding activity and the expressive intensity of phospho-STAT3 protein were lower in drug-resistant cell line. It can therefore be speculated that negative regulators of STAT signaling might be correlated with the decreased STAT3 activation in drug-resistant cell line.
In summary, our study demonstrates different activation of STAT3 signaling in these two human stomach adenocarcinoma cell lines, 5-fluorouracil resistant cell line and its parental cell line. The knowledge of STAT signaling pathway has proved to be of even broader benefit for illustrating the multi-drug resistance mechanisms of gastric cancer. The lower VEGF expression may be correlated with decreased STAT3 activation in drug-resistant cell line. Because the negative regulators of STAT signaling might play important roles in the control of tumor incidence and/or progression, further studies should be done in order to illustrate their regulation for tumors’ drug-resistance. The fact that STAT3 regulates the key angiogenic factor, VEGF, makes STAT3 a critical transcription factor in tumor development and progression. The parent cell line SGC7901, which is sensitive to the drugs of chemotherapy, has elevated constitutive activation of STAT3 and over-expression of VEGF, so targeting at STAT3 signaling may represent a novel approach to control the angiogenesis and growth of human gastric cancer.