Published online Nov 14, 2005. doi: 10.3748/wjg.v11.i42.6613
Revised: April 26, 2005
Accepted: April 30, 2005
Published online: November 14, 2005
AIM: To elucidate the molecular mechanisms of the inhibitory effects of IFN-α on tumor growth and metastasis in MHCC97 xenografts.
METHODS: Three thousand international units per milliliter of IFN-α-treated and -untreated MHCC97 cells were enrolled for gene expression analysis using cDNA microarray. The mRNA levels of several differentially expressed genes in cDNA microarray were further identified by Northern blot and RT-PCR.
RESULTS: A total of 190 differentially expressed genes including 151 IFN-α-repressed and 39 -stimulated genes or expressed sequence tags from 8 464 known human genes were found to be regulated by IFN-α in MHCC97. With a few exceptions, mRNA levels of the selected genes in RT-PCR and Northern blot were in good agreement with those in cDNA microarray.
CONCLUSION: IFN-α might exert its complicated anti-tumor effects on MHCC97 xenografts by regulating the expression of functional genes involved in cell metabolism, proliferation, morphogenesis, angiogenesis, and signaling.
- Citation: Wu WZ, Sun HC, Wang L, Chen J, Liu KD, Tang ZY. Modulation of gene expression in MHCC97 cells by interferon alpha. World J Gastroenterol 2005; 11(42): 6613-6619
- URL: https://www.wjgnet.com/1007-9327/full/v11/i42/6613.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i42.6613
Human hepatocellular carcinoma (HCC) is one of the most prevalent malignancies in China. Patients with HCC often die of tumor metastasis and recurrence even after curative resection. Recently, a metastatic human HCC model in nude mice (LCI-D20) and a series of HCC cell lines (MHCC97, MHCC97-H, MHCC97-L) with different metastatic potentials derived from LCI-D20 have been established in our institute[1,2]. Using this model, IFN-α significantly inhibits tumor growth and metastasis of MHCC97 xenografts has been found[3-5]. However, the underlying molecular mechanisms are still unclear.
IFN-α is a multifunctional cytokine capable of inter-fering with viral infection, inhibiting cell proliferation, regulating cell differentiation, as well as modulating immune response[6-9]. It is well known that these pleiotropic effects of IFN-α are mediated primarily through the tran-scriptional regulation of many different functional genes. Thanks to the rapid progress in human genetic projects; many functional human genes and expressed sequence tags (ESTs) are identified and released, which make us possible to use cDNA microarray to survey IFN-α-modulated genes in MHCC97 cells. In this study, we identified 190 differentially expressed genes from 8 464 known human genes, which might mediate various biological functions of IFN-α. These data provide us useful clues for further studying the anti-tumor mechanisms of IFN-α and finding the IFN-α mimics for HCC therapy.
MHCC97, a metastatic HCC cell line derived from LCI-D20 xenografts, was cultured in high glucose Dulbecco’s modified Eagle’s medium (Gibco-BRL, NY, USA) supplemented with 10% fetal calf serum (Hyclone, UT, USA), 100 U/mL penicillin and 100 μg/mL streptomycin in 20-cm2 tissue culture flasks. Cells were grown at 37 °C in a humidified atmosphere of 50 mL/L CO2 and passaged every 3 d.
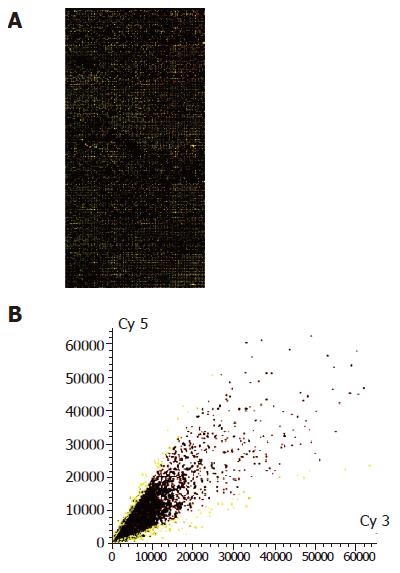
A total of 8 464 cDNAs of known human genes (United Gene Holding, Ltd, Shanghai) were amplified by polymerase chain reaction (PCR) using universal primers and spotted onto silylated slides (CEL Associates, Houston, TX, USA) using a Cartesian PixSys 7500 motion control robot (Cartesian Tech, Irvine, CA, USA) fitted with ChipMaker micro-spotting technology (TeleChem, Sunnyvale, CA, USA). After being hydrated, dried, cross linked and washed, the microarray was ready for use. Total RNA was isolated from IFN-α-treated and untreated (3 000 IU/mL, 16 h) cells using TRIzol (Gibco-BRL). cDNA probes were prepared by reverse transcription and purified according to the methods described by Schena et al[10]. Then equal amount of cDNA from IFN-α-untreated and treated MHCC97 cells was labeled with Cy3-dUTP and Cy5-dUTP, respectively. The mixed Cy3/Cy5 probes were purified and dissolved in 20 μL of hybridization solution (0.75 mol/L NaCl, 0.075 mol/L sodium citrate, 0.4% SDS, 50% formamide, 0.1% Ficoll, 0.1% polyvinylpyrrolidone and 0.1% BSA). Microarrays were pre-hybridized with 0.5 mg/mL salmon sperm DNA at 42 °C for 6 h. After being extensively washed, the denatured (95 °C, 5 min) fluorescent-labeled probe mixture was applied onto the pre-hybridized chips and further hybridized at 42 °C for 15-17 h under a cover glass. Subsequently, chips were sequentially washed for 10 min at 60 °C with 2×SSC+0.2% SDS, 0.1×SSC+0.2% SDS and 0.1×SSC solutions and dried at room temperature (1×SSC: 150 mmol/L NaCl, 15 mmol/L sodium citrate). Both Cy3 and Cy5 fluorescent signals of hybridized chips were scanned by ScanArray 4000 (GSI Lumonics, MA, USA) and analyzed using Genepix Pro 3.0 software (BioDiscovery Inc., CA, USA). To minimize artifacts arising from low expression, only genes whose Cy3 and Cy5 fluorescent intensities were both over 200 counts, or genes whose Cy3 or Cy5 fluorescent intensity was over 800 were selected for calculating the normalization cofactor (ln(Cy5/Cy3)). Genes were identified as differentially expressed, if the ratio of Cy5/(Cy3×normalization cofactor) (Cy5/Cy3*) was more than 2 or less than 0.5.
MHCC97 cells (106) cultured in 20-cm2 flasks were treated with 3 000 IU/mL IFN-α (Roche, Shanghai) for 0 or 16 h, and total RNA was extracted (RNeasy Mini Kit, QIAGEN Inc., CA, USA). One microgram RNA was used to set-up reverse transcription reactions (Gibco-BRL, NY, USA). Nine differentially expressed genes identified by cDNA microarray were selected for analysis by semi-quantitative PCR. Appropriate primers were designed using Primer3 software (http://www-genome.wi.mit.edu). γ-Actin was used as an internal standard. PCR reaction conditions and primer sequences are summarized in Table 1.
| Category | Gene | Sense and antisense primers | Annealing(°C) | Cycles | Size(bp) |
| Cytoskeletal gene | Neutral calponin | 5’-TGGCACCAGCTAGAAAACCT-3’; 5’-CAGGGACATGGAGGAGTTGT-3’ | 56 | 26 | 498 |
| Proliferative gene | hMCM2 | 5’-ACCGAGACAATGACCTACGG-3’; 5’-CTAGCTGTCTGCCCCTTGTC-3’ | 56 | 30 | 382 |
| Angiogenic gene | VEGF165 receptor | 5’-GAAGCACCGAGAGAACAAGG-3; 5’-CACCTGTGAGCTGGAAGTCA-3’ | 56 | 30 | 359 |
| IFN-α-induced genes | 9-27 | 5’-TTGGTCCCTGGCTAATTCAC-3’; 5’-ATGAGGATGCCCAGAATCAG-3’ | 53 | 35 | 491 |
| ISG-56 ku | 5’-AAAAGCCCACATTTGAGGTG-3’; 5’-GGCTGATATCTGGGTGCCTA-3’ | 54 | 30 | 451 | |
| MAPK pathway-related genes | ERK activator kinase (MEK2) | 5’-CGAAAGGATCTCAGAGCTGG-3’; 5’-GTGCTTCTCTCGGAGGTACG-3’ | 56 | 26 | 349 |
| G3BP2 | 5’-GCAGAACCTGTTTCTCTGCC-3’; 5’-CACCACCACCTCTGGTTTCT-3’ | 56 | 30 | 475 | |
| CHED | 5’-TCCTTGGCGAACTCTTCACT-3’; 5’-TGCCATAAAGGGAGATCTGG-3’ | 56 | 30 | 336 | |
| cAMP/PI3 pathway-related gene | Adenylyl cyclase | 5’-CCAGGAGCCTGAAGAATGAG-3’; 5’-GGCTTCTGAGCTCCAATCAC-3’ | 53 | 35 | 439 |
| Housekeeping gene | γ-Actin | 5’-ATGGAAGAAGAAATCGCCGC-3’; 5’-ACACGCAGCTCGTTGTAGAA-3’ | 55 | 25 | 287 |
Total RNA of 3 000 IU/mL IFN-α-treated or untreated MHCC97 cells was isolated as described above. Thirty microgram was separated by 1% agarose formaldehyde gel electrophoresis and transferred to a nylon membrane (Millipore, MA, USA) in 10×SSC by capillary blotting. The membrane was hybridized with the appropriate cDNA probe prepared from the human library of cDNA clones (Biostar Genechip Inc., Shanghai) and labeled with [α-32P]dCTP (Yahui Biomedical, Beijing) using random primer (Ambion Inc., Austin, TX, USA).
It is well known that the gene expression pattern of cells often varies with time and differentiation status and that cells derived from different individuals often have different genetic expression profiles. As a result, it is often difficult to extract useful information on the possible causes of phenotypic differences by comparing the genetic expression profiles of different cell lines. To minimize such complicated factors, we compared the gene expression profiles in 3 000 IU/mL IFN-α-treated and untreated (0 IU/mL) MHCC97 cells in two independent cDNA microarray analyses. We reasoned that such an internally consistent comparison might provide useful information on explaining the anti-tumor molecular mechanism of IFN-α in MHCC97 xenografts.
In 8 464 tested genes and ESTs, 190 genes were ide-ntified to be modulated by 3 000 IU/mL IFN-α treatment in MHCC97 cells. Among them the ex-pression of 151 genes was downregulated by IFN-α and the expression of 39 genes was upregulated by IFN-α. All differentially expressed genes are listed in Table 2 and the gene expression profiles obtained by cDNA microarray analysis are shown in Figure 1.
| Category | GenBank ID | Gene description | Cy5/Cy3* (average) |
| 2.1 Metabolism related genes | HUMCRTR | Creatine transporter | 0.251 |
| HSGAGMR | GARs-AIRs-GART | 0.289 | |
| HUM2OGDH | 2-Oxoglutarate dehydrogenase | 0.298 | |
| AF034544 | Delta7-sterol reductase | 0.318 | |
| HUMTK | Thymidine kinase | 0.333 | |
| HSU12778 | Acyl-CoA dehydrogenase | 0.341 | |
| HUMTHBP | Thyroid hormone-binding protein(p55) | 0.349 | |
| AF067127 | 7-Dehydrocholesterol reductase (DHCR) | 0.356 | |
| HSPRCOX | Pristanoyl-CoA oxidase | 0.364 | |
| AF035429 | Cytochrome oxidase subunit 1 | 0.372 | |
| AF070544 | Glucose transporter glycoprotein (SGLT) | 0.379 | |
| HSPFKLA | Liver-type1-phosphofructokianse (PFKL) | 0.392 | |
| HUMSHMT | Serine hydroxymethyltransferase 2 (SHMT2) | 0.407 | |
| HUMMGPHB | Brain glycogen phosphorylase | 0.413 | |
| HUMTCBA | Cytosolic thyroid hormone-binding protein (p58) | 0.415 | |
| D88152 | Acetyl-coenzyme A transporter | 0.451 | |
| HUMPKM2L | M2-type pyruvate kinase | 0.456 | |
| HSLDHBR | Lactate dehydrogenase B | 2.156 | |
| AF108211 | Inorganic pyrophosphatase | 2.25 | |
| HSCOXVII | Cytochrome C oxidase VII | 2.279 | |
| HUMCYCPSK | Cytochrome C (HS7) | 2.574 | |
| HUMDBI | Diazepam binding inhibitor | 2.628 | |
| 2.2 Proliferation, apoptosis and damaged DNA repairing related genes | HSATPBR | Na/K ATPase beta subunit | 0.208 |
| HUMP53T | Mutant p53 protein | 0.233 | |
| HSMITG | Mitochondrial DNA | 0.309 | |
| HSNUMAMR | Nuclear mitotic apparatus protein | 0.325 | |
| HSDNALIG3 | DNA ligase III | 0.34 | |
| G28520 | STS HSGC-31478 (homolog to Rad23a) | 0.341 | |
| AF096870 | Estrogen-responsive B box protein | 0.352 | |
| AF001609 | EXT like protein 3 | 0.367 | |
| AF015283 | Selenoprotein W | 0.369 | |
| AF011905 | Putative checkpoint control protein hRad1 | 0.398 | |
| HUMHMAM2 | Minichromosome maintenance 2 | 0.408 | |
| HUMRNAPII | RNA polymerase II 23 ku subunit | 0.408 | |
| AF007790 | Inversely correlated with estrogen receptor Expression (ICERE-1) | 0.413 | |
| HSU78310 | Pescadillo | 0.43 | |
| AF004162 | Nickel-specific induction protein (Cap43) | 0.434 | |
| HSU3298 | UV-damaged DNA binding factor | 0.437 | |
| HUMP1CDC47 | P1cdc47 | 0.442 | |
| HSU72649 | B cell translocation gene 2 | 0.444 | |
| AF031523 | bcl-xL/bcl-2 associated death promoter (BAD) | 0.481 | |
| AF132973 | CGI-39 (homolog to GRIM-19) | 2.079 | |
| 2.3 Morphogenesis, adhesion, and cytoskeleton | D38735 | Neutral calponin | 0.141 |
| AF006082 | Actin-related protein Arp2 | 0.197 | |
| U01244 | Fibulin 1D | 0.212 | |
| remodeling related genes | AF070593 | Beta tublin | 0.236 |
| HSU35622 | EWS-E1A-F chimeric protein | 0.255 | |
| AF049259 | Keratin 13 | 0.335 | |
| HSPRO4HY | Prolyl 4-hydoxylase beta | 0.337 | |
| HUMCN4GEL | Collagenase type IV | 0.36 | |
| AF005654 | Actin-binding double zinc-finger protein | 0.378 | |
| HSTEST | Testican | 0.379 | |
| HUMEPSURAN | Surface antigen | 0.389 | |
| AF004841 | CAM-related/down-regulated by oncogenes | 0.398 | |
| HUMGLBA | Co-beta-glucosidase | 0.402 | |
| HUMCA1XIA | Alpha-1 type XI collagen | 0.423 | |
| HUMMCPGV | Macrophage capping protein | 0.461 | |
| HUMNID | Nidogen | 0.497 | |
| HSTUMP | Translationally controlled tumor protein | 2.022 | |
| 2.4 Signal transmitting related genes | HUMEPHT2R | Protein tyrosine kinase (NET PTK) | 0.248 |
| HUMMEK2NF | ERK activator kinase (MEK2) | 0.271 | |
| HUMBADPTA | Beta-adaptin | 0.273 | |
| HUMP2A | Alpha-PR65 | 0.282 | |
| HUMHRGAA | rab GDI alpha | 0.285 | |
| AF053535 | ras-GAP/RNA binding protein G3BP2 | 0.296 | |
| HSRING3GE | RING 3 | 0.316 | |
| HSU45973 | Pt Ins (4,5) P(2) 5-phosphatase | 0.324 | |
| HSU07139 | Voltage-gated calcium channel beta | 0.329 | |
| HUMFTPB | Farnesyl-protein transferase beta | 0.345 | |
| HSU33053 | Lipid-activated protein kinase (PRK1) | 0.352 | |
| HUMHK1A | Calcium-ATPase (HK1) | 0.386 | |
| HSU66406 | EPH-related PTK receptor ligand LERK-8 | 0.386 | |
| HSPP15 | Placental protein 15 | 0.387 | |
| HSADCYCL | Adenylyl cyclase | 0.409 | |
| HUMCHED | cdc2-related protein kinase (CHED) | 0.412 | |
| AF093265 | Homer 3 | 0.415 | |
| HSU40282 | Integrin-linked kinase | 0.416 | |
| HUMGKAS | Stimulatory G protein | 0.416 | |
| HSU43939 | Nuclear transport factor 2 | 0.429 | |
| HUMCAK | Tyrosine protein kinase (CAK) | 0.439 | |
| HUMGNOS48 | Endothelial nitric oxide synthase | 0.443 | |
| HUMCDPKIV | Calmodulin-dependent protein kinase IV | 0.449 | |
| HSPKX1MR | Protein kinase, PKX1 | 0.469 | |
| D83760 | Mother against dpp (Mad) related protein | 0.472 | |
| HUMEGFGRBA | EGF receptor binding protein GRB2 | 0.481 | |
| HSU51004 | Protein kinase C inhibitor (PKCI-1) | 2.223 | |
| 2.5 Tumor angiogenesis related genes | HUMRNAMBPE | Golli-mbp | 0.236 |
| AF016050 | VEGF 165 receptor/neuropilin | 0.25 | |
| AF001307 | Aryl hydrocarbon receptor nuclear translocator | 0.27 | |
| HSU64791 | Golgi membrane sialoglycoprotein MG 160 | 0.355 | |
| HUMPTPRZ | Protein tyrosine phosphatase Zeta-polypeptide | 0.363 | |
| HSU28811 | Cysteine-rich FGFR (CFR1) | 0.414 | |
| HSU20758 | Osteopontin | 2.193 | |
| HUMTR107 | DNA-binding protein, TAXREB107 | 2.24 | |
| HUMNEPPON | Nephropontin | 2.413 | |
| 2.6 Transcriptional activity related genes | S66431 | Retinoblastoma binding protein 2 | 0.182 |
| HUMANT61K | Medium antigen-associated 61 ku protein | 0.183 | |
| HSU58197 | Interleukin enhancer binging factor 2 | 0.226 | |
| HSUBP | Upstream binding factor | 0.266 | |
| 4758315 | ets-related molecule, ETV5 | 0.267 | |
| AF099013 | Glucocorticoid modulatory element binding protein 1 | 0.309 | |
| HSU72621 | Lost on transformation 1(LOT1) | 0.313 | |
| HUMFOS | Oncogene protein, c-fos | 0.361 | |
| AB019524 | Nuclear receptor co-repressor | 0.369 | |
| HS14AGGRE | Conserved gene telomeric to alpha globin cluster | 0.398 | |
| HSU74667 | tat interactive protein (tip60) | 0.404 | |
| AF114816 | KRAB-zinc finger protein SZF1-1 | 0.406 | |
| HSU80456 | Drosophila single-minded, SIM2 | 0.409 | |
| AF117756 | TRAP 150 | 0.41 | |
| HSU15306 | Cysteine rich DNA binding protein NFX1 | 0.417 | |
| S57153 | Retinoblastoma binding protein 1 | 0.469 | |
| HUM56KDAPR | IEF SSP 9502 | 2.183 | |
| HUMTR107 | DNA binding protein. TAXREB 107 | 2.24 | |
| HUMMSS1 | Mammalian suppressor of sgv 1, MSS 1 | 2.313 | |
| 2.7 mRNA and protein processing, secretory, proteolysis related genes | HSU39412 | Alpha SNAP | 0.141 |
| HSU47927 | Isopeptidase T (ISOT) | 0.229 | |
| HSU72355 | hsp27 ERE-TATA bind protein, HET | 0.231 | |
| AF077039 | TIM17 homolog | 0.238 | |
| HUMHRH1 | RNA helicase, HRH1 | 0.251 | |
| AF206402 | U5 SnRNP 100 ku protein | 0.255 | |
| D85429 | Heat shock protein 40 | 0.344 | |
| HSU85946 | hSec 10p | 0.378 | |
| HSY10806 | Arginine methyltransferase | 0.412 | |
| AB002135 | Glycophosphatidylinositol anchor attachment 1 | 0.428 | |
| AB007510 | PRP8 protein | 0.436 | |
| HSU24105 | Coatomer protein (COPA) | 0.455 | |
| HSCANPX | Calpain-like protease (CANPX) | 0.456 | |
| HSRBPRL7A | Ribosomal protein L7 | 2.067 | |
| D89678 | A+U-rich element RNA-binding protein | 2.069 | |
| HSU14966 | Ribosomal protein L5 | 2.113 | |
| HSRPL31 | Ribosomal protein L31 | 2.142 | |
| HUMPSC9 | Proteasome subunit HC9 | 2.179 | |
| HSU26312 | Heterochromatin protein HP1 HS-gamma | 2.182 | |
| HUMRPS7A | Ribosomal protein S7 | 2.289 | |
| AF106622 | TIM17a | 2.312 | |
| HSUCEH3 | Ubiquitin-conjugated enzyme UbCH2 | 2.323 | |
| HUMRPS7A | Ribosomal protein S7 | 2.289 | |
| AF106622 | TIM17a | 2.312 | |
| HSUCEH3 | Ubiquitin-conjugated enzyme UbCH2 | 2.323 | |
| HUMRPS25 | Ribosomal protein S25 | 2.326 | |
| HUMRPSA3A | Ribosomal protein S3a | 2.328 | |
| HSRNASMG | Sm protein G | 2.334 | |
| HUMRPS18 | Ribosomal protein S18 | 2.341 | |
| HUMRP4SX | Ribosomal protein S4 isoform | 2.346 | |
| HUMPSC3 | Proteasome subunit HC3 | 2.368 | |
| HUMTCP20 | Chaperonin protein, TCP20 | 2.572 | |
| 4504522 | Chaperonin protein, hsp10 | 2.686 | |
| 2.8 Tumor antigen processing, anti-viral infection related genes | HUMSAPC1 | Cerebroside sulfate activator protein | 0.211 |
| AF077011 | Interleukin 16 | 0.23 | |
| AF057307 | Prosaposin | 0.26 | |
| HUMSIATA | Sialyltransferase | 0.26 | |
| AF055008 | Epithelin 1 and 2 | 0.363 | |
| HSU58766 | FX protein | 0.393 | |
| HUMOSF1 | OSF1 | 0.407 | |
| HSU46194 | RAGE 4 | 0.43 | |
| HSU18121 | 136 ku double-stranded RNA binding protein | 0.469 | |
| AF021315 | Reverse transcriptase | 0.483 | |
| S74095 | Preproenkephalin A | 2.115 | |
| HUM927A | Interferon inducible protein 9-27 | 2.356 | |
| HSIFI56R | Interferon inducible protein 56 ku | 3.829 | |
| HUMHCAMAP1 | Interferon inducible protein 44 ku | 4.03 | |
| 2.9 Genes with unknown biological functions | D50928 | KIAA0138 | 0.23 |
| AF132942 | CGI08 | 0.269 | |
| AB020677 | KIAA0870 | 0.271 | |
| AB011110 | KIAA0538 | 0.277 | |
| AB028956 | KIAA1033 | 0.28 | |
| HSU10362 | GB36b glycoprotein | 0.335 | |
| 4579277 | A homolog of proteasome regulatory S2 | 0.352 | |
| AB002356 | KIAA0358 | 0.371 | |
| 4505130 | A homolog of MCM3 | 0.371 | |
| AB029020 | KIAA1097 | 0.381 | |
| HS130N43 | 0.383 | ||
| HSU66406 | Eplg8 | 0.386 | |
| HSNIPSNA1 | NIPSNAP1 protein | 0.391 | |
| AB002378 | KIAA0380 | 0.405 | |
| HSU90907 | Regulatory subunit of P55 PIK | 0.407 | |
| AB208959 | KIAA1036 | 0.414 | |
| AB020658 | KIAA0851 | 0.416 | |
| AF035282 | 0.416 | ||
| AF000136 | 0.419 | ||
| HUMORFFA | KIAA0120 | 0.424 | |
| D13699 | KIAA0019 | 0.43 | |
| HUMORFB1 | KIAA0123 | 0.432 | |
| AF151830 | CGI72 | 0.436 | |
| AB007900 | KIAA0440 | 0.437 | |
| AB014595 | KIAA0695 | 0.439 | |
| HSM800064 | 0.439 | ||
| HUMORFA04 | KIAA0115 | 0.457 | |
| HSU79287 | 0.462 | ||
| AF007149 | 0.473 | ||
| AF007135 | 2.147 | ||
| AF151875 | CGI117 | 2.184 | |
| AF151857 | CGI99 | 2.326 | |
| HUMRSC508 | KIAA0020 | 2.45 |
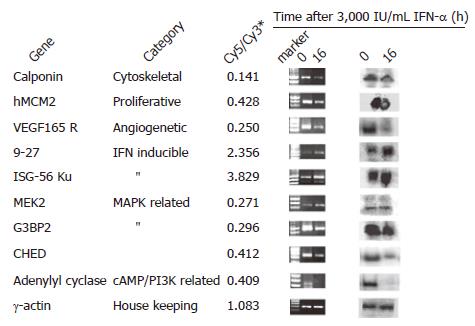
To validate the results of cDNA microarray, we selected nine genes whose expressions were clearly altered by IFN-α and evaluated their expressions by PCR and Northern blot. We enrolled IFN-α-regulated genes and found that the results were consistent with the previous reports[11,12].
For PCR analysis, we synthesized primers as indicated in Table 1 and performed semi-quantitative RT-PCR as outlined under “Materials and methods” after treatment of MHCC97 cells with 3 000 IU/mL IFN-α for 0 or 16 h. The transcription patterns of the same genes were also analyzed by Northern blot. Among the nine selected genes, seven downregulated genes were proved by cDNA microarray, six by RT-PCR and five by Northern blot analysis. Two stimulated genes, ISG-56 ku and 9-27 were proved by cDNA microarray, RT-PCR and Northern blot analysis. ERK activator kinase (MEK2), one repressed gene in cDNA microarray, was not changed in RT-PCR or Northern blot analysis. Thus, with a few exceptions, the results of RT-PCR and Northern blot were in good agreement with those of cDNA microarray analysis (Figure 2).
cDNA microarray is a useful technique for rapid screening of gene expressions in cells, although the results need to be further confirmed by other molecular methods. Using this method, we found 211 hybrid dots, whose Cy5/Cy3* ratio was either more than 2 or less than 0.5 in IFN-α-treated MHCC97. Blasting the cDNA sequences in public database showed that these dots represented 190 different human genes or ESTs due to the redundant hybrids. Based on the results of RT-PCR and Northern blot, we believe that our cDNA microarray data are reliable. These differentially expressed genes might mediate the multiple biological functions of IFN-α directly or indirectly in MHCC97. We have artificially categorized these genes into nine functional clusters (Table 2).
IFN-α might interfere with cellular metabolisms by downregulating metabolic gene expression. In detail, IFN-α can inhibit glycolysis, glycogen degradation, gluconeogenesis as well as creatine or glucose tran-sportation by repressing the expressions of liver-type phosphofructokinase (hPFKL), M2-type pyruvate kinase, brain glycogen phosphorylase, 2-oxoglutarate de-hydrogenase, glucose transporter glycoprotein (SGLT) and cytosolic thyroid hormone-binding protein[13]. IFN-α can also inhibit lipolysis by reducing the expression of delta7-sterol reductase and pristanoyl-CoA oxidase, two key enzymes in lipid metabolism[14,15]. In addition, IFN-α reduces purine and pyridine biosynthesis by repressing the expression of GARs-AIRs-GART and serine hydro-xymethyltransferase 2 (SHMT2). All these indicate that IFN-α-treated MHCC97 can result in lower ATP production and DNA synthesis, and slow down cell proliferation.
Many proliferation-, apoptosis- and cell cycle-regulating genes are modulated by IFN-α in MHCC97. Downregulating the expression of mutant p53, mito-chondrial DNA, nuclear mitotic apparatus protein (NuMA), and RNA polymerase II 23 ku subunit (polR2) might cause cell cycle arrest[16,17]. Downregulating the expression of DNA ligase III, hRad1, minichromosome maintenance 2 (hMCM2) as well as UV-damaged DNA binding factor might hinder damaged DNA repairing[18,19]. Stimulating retinoid-IFN-induced mortality 19 (GRIM-19) expression might promote IFN-α-induced apoptosis[20].
Several genes functionally related to cell morphogenesis, adhesion, and cytoskeleton remodeling are also modulated by IFN-α in MHCC97. For example, decreasing the expression of calponin, actin-related protein 2 (Arp2), fibulin 1D, beta-tublin and epidermal surface antigen (ESA), etc., might damage mitotic spindle formation and might interfere with actin-based cell motility, migration, adhesion and morphogenesis[21-24]. Reducing the expression of prolyl 4-hydroxylase beta, a key enzyme in collagen biosynthesis and type IV collagenase, a tumor-derived extracellular matrix metalloproteases might block tumor invasion and metastasis. Although most genes in this category were first identified as IFN-α regulating genes, their roles in mediating IFN-α functions need to be further studied.
In this study, we found that many genes functionally related to signal transmitting were affected by IFN-α in MHCC97. By repressing the expressions of discoidin domain receptor, integrin-linked kinase, EPH-related tyrosine kinase (EPT2) and MEK2, etc., IFN-α might block cellular signaling initiated by tyrosine-kinase receptors[25,26]. By modulating the expressions of Rab GDI, Ras-related GTP-binding proteins and farnesyl-protein transferase and nuclear transport factor (NTF2) and G3BP2, a Ras-GAP/RNA binding protein, IFN-α might interfere with GTP/GDP exchange and nuclear import, thus influencing the recycles and activities of ras and its homologs[27-29]. By attenuating the expressions of adenylyl cyclase (AC) and phosphatidylinositol 4,5-bisphosphate 5-phosphatase (PtdIns (4,5)P(2)5- phospharase), a catalyzer of pho-sphatidylinositol 4,5-bisphosphate and PRK1, IFN-α might decrease inositol polyphosphate levels in cytosol and might inhibit the serine/threonine-kinase activities through cAMP/ PI3P signal pathway[30,31]. All these changes might exert inhibitory effects of IFN-α on MAPK and PI3K signaling. In addition, other signaling pathways such as Ca(2+), NO and TGFβ/hMAD-dependent signaling pathways are suppressed by IFN-α as well[32,33]. Plausibly Jak/STATs pathway, the most important IFN-α signaling pathway, is confirmed not to be regulated in IFN-α-treated MHCC97. The deficient expression of p48 (ISGF3γ) in this cell line may be the possible mechanism for the non-response of IFN-α priming via Jak/STATs pathway (data not shown).
In this study, we found that many angiogenic-related genes were regulated by IFN-α. By attenuating the expressions of Golli-MBP[34], VEGF 165 receptor and aryl hydrocarbon receptor nuclear translocator (ARNT)[35] as well as Golgi membrane sialoglycoprotein MG 160, a bFGF binding protein and cysteine-rich FGF receptor (CFR-1)[36], IFN-α may destroy the balance between pro- and anti-angiogenic factors and exert its inhibitory effects on tumor angiogenesis.
It is well known that cells usually respond to various stimuli by rapidly shifting the functions of transcriptional factors. Using this strategy, IFN-α might impose its anti-proliferative functions and hormone response by fluctuating the expression of several transcriptional factors or their cofactors such as retinoblastoma binding protein2 (RBP2), interleukin enhancer binding factor 2, lost on transformation 1 (LOT1) and KRAB-zinc finger protein (SZF1)[37-40].
In addition, IFN-α might hinder with mRNA/rRNA spicing and maturation by downregulating RNA helicase (HRH1), U5 snRNP[41] and affect protein transportation, secretion and proteolysis by downregulating alpha SNAP, GPAA1, hSec10p, hsp40 and isopeptidase T, a putative molecular in ubiquitin–proteasome pathway[42-44]. Meanwhile IFN-α might evoke anti-viral or tumor immune response by upregulating 9-27, 56 ku protein and p44 expressions.
Except for functionally definite genes, many ESTs with unknown functions were identified as IFN-α-regulated genes in our study (Table 2). In conclusion, cDNA microarray is a useful, rapid method for screening transcriptome of cells and potentially paves a way for elucidating IFN-α effects on tumor growth and metastasis.
We thank Shanghai Biostar Genechip Inc. for cDNA microarray service.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Sun FX, Tang ZY, Lui KD, Ye SL, Xue Q, Gao DM, Ma ZC. Establishment of a metastatic model of human hepatocellular carcinoma in nude mice via orthotopic implantation of histologically intact tissues. Int J Cancer. 1996;66:239-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 2. | Tian J, Tang ZY, Ye SL, Liu YK, Lin ZY, Chen J, Xue Q. New human hepatocellular carcinoma (HCC) cell line with highly metastatic potential (MHCC97) and its expressions of the factors associated with metastasis. Br J Cancer. 1999;81:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 224] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 3. | Wang L, Tang ZY, Qin LX, Wu XF, Sun HC, Xue Q, Ye SL. High-dose and long-term therapy with interferon-alfa inhibits tumor growth and recurrence in nude mice bearing human hepatocellular carcinoma xenografts with high metastatic potential. Hepatology. 2000;32:43-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 4. | Wu WZ, Sun HC, Gao YQ, Li Y, Wang L, Zhou K, Liu KD, Iliakis G, Tang ZY. Reduction in p48-ISGFgamma levels confers resistance to interferon-alpha2a in MHCC97 cells. Oncology. 2004;67:428-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Wu WZ, Sun HC, Shen YF, Chen J, Wang L, Tang ZY, Iliakis G, Liu KD. Interferon alpha 2a down-regulates VEGF expression through PI3 kinase and MAP kinase signaling pathways. J Cancer Res Clin Oncol. 2005;131:169-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Tough DF, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947-1950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 866] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 7. | Albini A, Marchisone C, Del Grosso F, Benelli R, Masiello L, Tacchetti C, Bono M, Ferrantini M, Rozera C, Truini M. Inhibition of angiogenesis and vascular tumor growth by interferon-producing cells: A gene therapy approach. Am J Pathol. 2000;156:1381-1393. [RCA] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Slaton JW, Perrotte P, Inoue K, Dinney CP, Fidler IJ. Interferon-alpha-mediated down-regulation of angiogenesis-related genes and therapy of bladder cancer are dependent on optimization of biological dose and schedule. Clin Cancer Res. 1999;5:2726-2734. [PubMed] |
| 9. | Hong YK, Chung DS, Joe YA, Yang YJ, Kim KM, Park YS, Yung WK, Kang JK. Efficient inhibition of in vivo human malignant glioma growth and angiogenesis by interferon-beta treatment at early stage of tumor development. Clin Cancer Res. 2000;6:3354-3360. [PubMed] |
| 10. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5102] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 11. | Elco CP, Guenther JM, Williams BR, Sen GC. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol. 2005;79:3920-3929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Martensen PM, Justesen J. Small ISGs coming forward. J Interferon Cytokine Res. 2004;24:1-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 13. | Ishikawa N, Oguri T, Isobe T, Fujitaka K, Kohno N. SGLT gene expression in primary lung cancers and their metastatic lesions. Jpn J Cancer Res. 2001;92:874-879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Witsch-Baumgartner M, Löffler J, Utermann G. Mutations in the human DHCR7 gene. Hum Mutat. 2001;17:172-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Jia Y, Qi C, Zhang Z, Hashimoto T, Rao MS, Huyghe S, Suzuki Y, Van Veldhoven PP, Baes M, Reddy JK. Overexpression of peroxisome proliferator-activated receptor-alpha (PPARalpha)-regulated genes in liver in the absence of peroxisome proliferation in mice deficient in both L- and D-forms of enoyl-CoA hydratase/dehydrogenase enzymes of peroxisomal beta-oxidation system. J Biol Chem. 2003;278:47232-47239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038-4043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 193] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Taimen P, Viljamaa M, Kallajoki M. Preferential expression of NuMA in the nuclei of proliferating cells. Exp Cell Res. 2000;256:140-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Maiorano D, Lemaître JM, Méchali M. Stepwise regulated chromatin assembly of MCM2-7 proteins. J Biol Chem. 2000;275:8426-8431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187-3196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 163] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 20. | Chidambaram NV, Angell JE, Ling W, Hofmann ER, Kalvakolanu DV. Chromosomal localization of human GRIM-19, a novel IFN-beta and retinoic acid-activated regulator of cell death. J Interferon Cytokine Res. 2000;20:661-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Curtis M, Nikolopoulos SN, Turner CE. Actopaxin is phosphorylated during mitosis and is a substrate for cyclin B1/cdc2 kinase. Biochem J. 2002;363:233-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 241] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 23. | Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997;138:575-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 24. | Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793-13802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 466] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 25. | Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 859] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 26. | Tang XX, Biegel JA, Nycum LM, Yoshioka A, Brodeur GM, Pleasure DE, Ikegaki N. cDNA cloning, molecular characterization, and chromosomal localization of NET(EPHT2), a human EPH-related receptor protein-tyrosine kinase gene preferentially expressed in brain. Genomics. 1995;29:426-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Ishizaki H, Miyoshi J, Kamiya H, Togawa A, Tanaka M, Sasaki T, Endo K, Mizoguchi A, Ozawa S, Takai Y. Role of rab GDP dissociation inhibitor alpha in regulating plasticity of hippocampal neurotransmission. Proc Natl Acad Sci USA. 2000;97:11587-11592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Prigent M, Barlat I, Langen H, Dargemont C. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000;275:36441-36449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Brassard DL, English JM, Malkowski M, Kirschmeier P, Nagabhushan TL, Bishop WR. Inhibitors of farnesyl protein transferase and MEK1,2 induce apoptosis in fibroblasts transformed with farnesylated but not geranylgeranylated H-Ras. Exp Cell Res. 2002;273:138-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 694] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 31. | Di Pasquale G, Stacey SN. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J Virol. 1998;72:7916-7925. [PubMed] |
| 32. | Tamura N, Tai Y, Sugimoto K, Kobayashi R, Konishi R, Nishioka M, Masaki T, Nagahata S, Tokuda M. Enhanced expression and activation of Ca(2+)/calmodulin-dependent protein kinase IV in hepatocellular carcinoma. Cancer. 2000;89:1910-1916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 33. | Mostert V, Dreher I, Köhrle J, Wolff S, Abel J. Modulation of selenoprotein P expression by TGF-beta(1) is mediated by Smad proteins. Biofactors. 2001;14:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 34. | Baron P, Constantin G, Meda L, Scarpini E, Scarlato G, Trinchieri G, Monastra G, Rossi F, Cassatella MA. Cultured human monocytes release proinflammatory cytokines in response to myelin basic protein. Neurosci Lett. 1998;252:151-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 35. | Onita T, Ji PG, Xuan JW, Sakai H, Kanetake H, Maxwell PH, Fong GH, Gabril MY, Moussa M, Chin JL. Hypoxia-induced, perinecrotic expression of endothelial Per-ARNT-Sim domain protein-1/hypoxia-inducible factor-2alpha correlates with tumor progression, vascularization, and focal macrophage infiltration in bladder cancer. Clin Cancer Res. 2002;8:471-480. [PubMed] |
| 36. | Shen B, Arese M, Gualandris A, Rifkin DB. Intracellular association of FGF-2 with the ribosomal protein L6/TAXREB107. Biochem Biophys Res Commun. 1998;252:524-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 37. | López-Fernández LA, Párraga M, del Mazo J. Ilf2 is regulated during meiosis and associated to transcriptionally active chromatin. Mech Dev. 2002;111:153-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 38. | Cao X, Südhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 895] [Cited by in RCA: 938] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 39. | Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJ. Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000;275:18000-18010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 40. | Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J Biol Chem. 2002;277:10236-10243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 41. | Teigelkamp S, Mundt C, Achsel T, Will CL, Lührmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313-1326. [PubMed] |
| 42. | Moro F, Sirrenberg C, Schneider HC, Neupert W, Brunner M. The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 1999;18:3667-3675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 73] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Hiroi Y, Chen R, Sawa H, Hosoda T, Kudoh S, Kobayashi Y, Aburatani H, Nagashima K, Nagai R, Yazaki Y. Cloning of murine glycosyl phosphatidylinositol anchor attachment protein, GPAA1. Am J Physiol Cell Physiol. 2000;279:C205-C212. [PubMed] |
| 44. | Hernández MP, Chadli A, Toft DO. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J Biol Chem. 2002;277:11873-11881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 103] [Article Influence: 4.5] [Reference Citation Analysis (0)] |