Published online Nov 7, 2005. doi: 10.3748/wjg.v11.i41.6429
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: November 7, 2005
AIM: To investigate the inhibitory effect of VP22 fusion protein-based dominant negative (DN) mutant on Hepatitis Bvrus (HBV) replication.
METHODS: Full-length or truncated fragment of VP22 was fused to C terminal of HBV core protein (HBc), and subcloned into pcDNA3.1 (-) vector, yielding eukaryotic expression plasmids of DN mutant. After transfection into HepG2.2.15 cells, the expression of DN mutant was identified by immunofluorescence staining. The inhibitory effect of DN mutant on HBV replication was indexed as the supernatant HBsAg concentration determined by RIA and HBV-DNA content by fluorescent quantification-PCR (FQ-PCR). Meanwhile, metabolism of HepG2.2.15 cells was evaluated by MTT colorimetry.
RESULTS: VP22-based DN mutants and its truncated fragment were expressed in HepG2.2.15 cells, and had no toxic effect on host cells. DN mutants could inhibit HBV replication and the transduction ability of mutant-bearing protein had a stronger inhibitory effect on HBV replication. DN mutants with full length of VP22 had the strongest inhibitory effect on HBV replication, reducing the HBsAg concentration by 81.94%, and the HBV-DNA content by 72.30%. MTT assay suggested that there were no significant differences in cell metabolic activity between the groups.
CONCLUSION: VP22-based DN mutant can inhibit HBV replication effectively.
- Citation: Yi J, Gong WD, Wang L, Ling R, Chen JH, Yun J. VP22 fusion protein-based dominant negative mutant can inhibit hepatitis B virus replication. World J Gastroenterol 2005; 11(41): 6429-6432
- URL: https://www.wjgnet.com/1007-9327/full/v11/i41/6429.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i41.6429
At present, about 3.5 hundred million people are infected with HBV, and about 1 million people die of Hepatitis Bvrus (HBV) infection or related diseases each year, ranking 9th in disease deaths[1-4]. There are no effective therapies for HBV infection. Although about 40% HBV-infected patients responded to interferon, patients in our country have a poorer response to it. Other antiviral agents, such as nucleoside/nucleotide analogs[5], are quite effective inhibitors of HBV replication[6-9]. However, the rapid development of drug resistance remains a growing concern. Gene therapy provides a novel idea for the treatment of HBV infection, can inhibit replication of HBV at gene level and remove HBV from host cells[10-12]. Expression of dominant negative (DN) mutant in liver cells is one of the most important strategies[13-17].
In 1997, Elliott and O’Hare[18] found that VP22, a structural protein (and its fusion protein) of herpes simplex virus type-1 (HSV-1), can enter into the cells from the medium, and translocate between cells in a contact-independent manner. Here, VP22 was fused to HBV core protein (HBc) and a fusion DN mutant was constructed to enhance the antiviral effect of the DN mutant by utilizing the transduction ability of VP22 protein.
Plasmids pVP22/myc-His2 and pcDNA3.1(-) were purchased from Invitrogen Co. EBO-HBV, in which 1.3-folds of HBV genome was cloned, and stored in our laboratory. Restriction enzymes and ligations were purchased from TaKaRa Biotech Co., Ltd; mouse anti-HBc or c-myc antibodies were purchased from Santa Cruz Co. FITC-labeled sheep anti-mouse IgG was purchased from BioStar Biotech Co., Ltd. DMEM, fetal bovine serum, and Lipofectamine 2000 were purchased from GIBCO Co. Solid phase radio-immunoassay kit was purchased from Beimian Dongya Biotech Institute, Beijing.
All the oligonucleotides and primers were synthesized by TaKaRa Biotech Co., Ltd. The underlined sequences indicate restriction enzyme (right) sites, respectively, which were added to the oligonucleotide primers for subsequent cloning of amplified DNA. The capital letters indicate start or stop codon.
HBc1: 5’-gcgcggtaccATGgacatcgaccctt-3’ (KpnI)
HBc2: 5’-gcgcctcgagTCAggatccacattgaggttccc-3’ (XhoI, BamHI)
US: 5’-gcccggatccatgacctctcgctccgtg-3’ (BamHI)
UR: 5’-ccccagatctatcgggactcgccatacc-3’ (BglII)
DS: 5’-gccgagatctgacgcggccacggcg-3’ (BglII)
DR: 5’-gggcctcgagTTAcagctaatcctcttctgag-3’ (XhoI)
c-myc1:5’-gcccggatccgaacaaaaactcatctcagaagaggatctgTAAc
tcgaggggc-3’ (BamHI, XhoI)
c-myc2:5’-gggcctcgagTTAcagatcctcttctgagatgagtttttgttcggatc
cgccc-3’ (XhoI, BamHI)
HepG2.2.15 cells integrated with full-length HBV gene[19-21] were cultured in DMEM containing 150 mL/L fetal bovine serum at 37 °C, in 50 mL/L CO2. G418 was added to screen cells at the final concentration of 100 mg/L. The media were freshened once in every two days and the cells were passaged every six days.
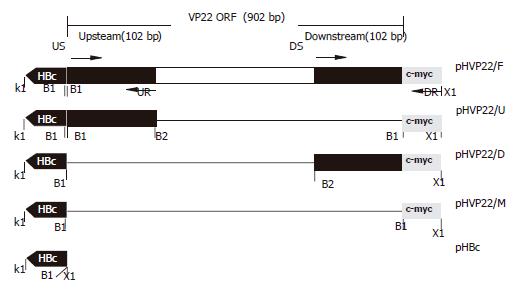
PCR products, amplified from EBO-HBV using HBc1/HBc2 as primers, were cloned into pcDNA3.1(-) after being digested by KpnI/XhoI, yielding the vector pHBc (Figure 1).
C-myc1/c-myc2 were dissolved at a concentration of 0.1 g/L and annealed by heating at 100 °C for 5 min, then slowly cooling to room temperature to form double-stranded c-myc digested by BamHI/XhoI and cloned into pHBc digested by the same restrictions to produce plasmid pHVP22/M.
The fragment including full-length of VP22 and c-myc epitope was amplified from pVP22/myc-His2 plasmid (primers US/DR). PCR products were digested by BamHI/XhoI and inserted into pHBc to produce pHVP22/F.
The upstream of VP22-coding sequence 102 bp amplified from pVP22/myc-His2 (primers US/UR) was digested by BamHI/BglII and ligated into BamHI site of pHVP22/M restricted with BamHI and dephosphorized. The resulting plasmid bearing directional insertion was called pHVP22/U.
The downstream of VP22-coding sequence 102 bp was amplified from pVP22/myc-His2, and PCR products were inserted into pHBc after BamHI/XhoI digestion to produce plasmid pHVP22/D.
Transfections were performed as described by the provider of Lipofectamine 2000 (Gibco’s handbook). Cells (4×108 L) were added to a 24-well plate (500 L/well) in which a coverslide was placed in advance. The transfection experiment was divided into seven groups and carried out in triplicate: pHVP22/F, pHVP22/U, pHVP22/D, pHVP22/M, pHBc, pcDNA3.1(-) and MOCK transfection.
After transfection, cells were immediately washed with sterilized PBS (4 °C, pH 8.0), fixed in 20 g/L paraformaldehyde and 1 g/L Triton X-100 diluted in PBS, and put on ice for 30 min. Cells were washed thrice with cold PBS. Non-specific epitopes were blocked with 10 g/L BSA for 10 min at 42 °C. After being washed with cold PBS, cells were incubated with mouse anti-HBc or c-myc mAb (1:500) for 15 min at 42 °C, further incubated with rabbit anti-mouse IgG labeled with FITC (1:1 000). The coverslides were mounted on slides using 50 mL/L glycerol/PBS and observed by fluorescence microscopy.
Forty-eight hours after transfection, HBsAg concentration was determined by RIA kit (completed by Nuclear Medicine Department of Xijing Hospital), and HBV-DNA content was quantified by FQ-PCR. The data obtained were analyzed by SPSS software.
The effect of transgene expression on host cells was evaluated by MTT colorimetry.
To enhance the antiviral effect of HBc DN mutant, both the full length of VP22 and the truncated version (102 bp upstream or downstream) were fused to C terminals of HBc, respectively (pHVP22/F, pHVP22/U, pHVP22/D), and the effects of anti-HBV replication were detected. All plasmids used here were confirmed by sequencing, which was completed by GeneCore Company (Shanghai, China).
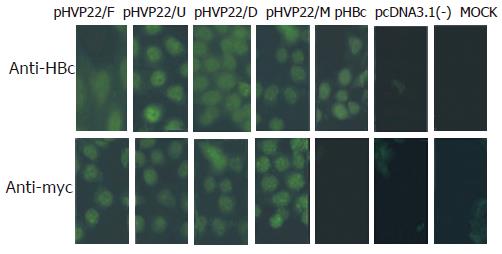
To identify the expression of transgenes in HepG2.2.15 cells, indirect immunofluorescence was performed using antibody against HBc or c-myc. No fluorescence was found in pcDNA3.1(-) and MOCK transfection, the same results were obtained in pHBc, pcDNA3.1(-) and MOCK transfection. Strong fluorescence was detected in other transfections. The results suggested that transgenes were successfully expressed in HepG2.2.15 cells and recognized by the corresponding antibodies (Figure 2).
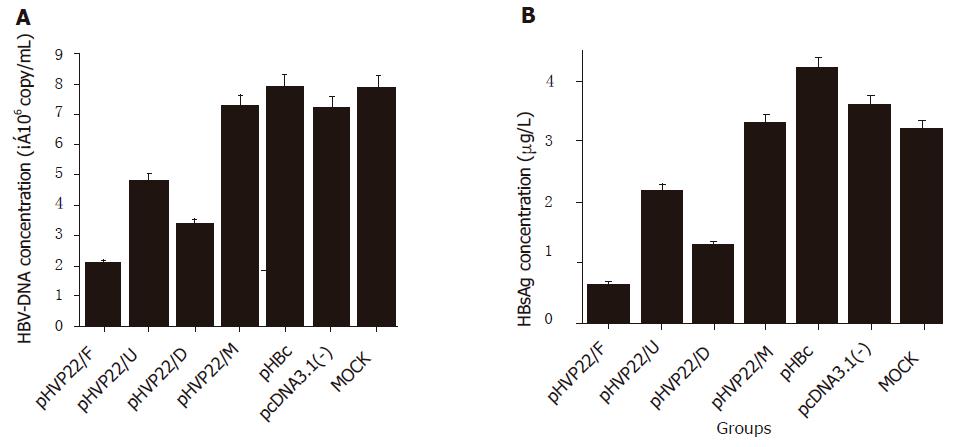
To investigate the antiviral activity of VP22 fusion DN mutant on HBV replication, the inhibitory effect was determined by HBsAg concentration and HBV-DNA content in the supernatant of HepG2.2.15 cell culture. When compared to MOCK transfection, HBsAg concentrations in pHVP22/F, pHVP22/U, and pHVP22/D transfections decreased by 81.94%, 38.88%, and 63.89%, respectively (Figure 3A, P<0.05), and HBV-DNA content by 72.30%, 29.60%, 46.42%, respectively (Figure 3B, P<0.05), indicating that VP22 fusion DN mutant could inhibit HBV replication effectively. Regardless of taking HBsAg concentration or HBV-DNA content as an index, pHVP22/F, pHVP22/U, and pHVP22/D showed a significant difference between the groups (P<0.05). DN mutant with transduction domains (pHVP22/F and pHVP22/D) had a stronger antiviral activity than that without the domain (pHVP22/U, P<0.05). When compared to DN mutant with full length of VP22 (pHVP22/F), DN mutant with the transduction domain (pHVP22/D) showed a weaker antiviral activity (P<0.05). There was no significant difference between DN mutant based on c-myc epitope (pHVP22/M) and MOCK transfection (P>0.05), suggesting that pHVP22/M could not inhibit HBV replication.
After 48-h incubation, the morphology of cells was observed under inverted microscope and no discernable difference was found between groups. MTT assay showed no significant differences between groups (P>0.05), suggesting that the expression of DN mutants had no effect on the growth of host cells (Table 1).
| Group | A490nm | P1 |
| pHVP22/F | 0.425 ± 0.065 | >0.05 |
| pHVP22/U | 0.465 ± 0.050 | >0.05 |
| pHVP22/D | 0.410 ± 0.075 | >0.05 |
| pHVP22/M | 0.438 ± 0.042 | >0.05 |
| pHBc | 0.413 ± 0.063 | >0.05 |
| pcDNA3.1(–) | 0.430 ± 0.065 | >0.05 |
| MOCK | 0.423 ± 0.058 | >0.05 |
HBV infection is an important health problem worldwide[14], and the investigation about HBV therapy is a long-standing focus. The development of genetic engineering facilitates the role of gene therapy in anti-HBV therapy[13,14] and DN mutant is one of the important strategies[15-17]. After introduction of mutant gene into the infected cells, the activity of wild-type viral gene is inhibited by the mutant version in a competitive manner, leading to the inhibition of virus replication[22,23]. DN mutant, as a preferential strategy, takes protein as the effector and avoids the problem of HBV mutation, which is one of the obstacles when DNA is used as an effector. But insufficient therapeutic molecules can reach the VP22 protein, bearing protein-transduction ability, to enhance the DN mutant effect and complement the shortage of therapeutic molecules in target cells.
As a structural protein of HSV-1, VP22 (or its fusion protein) has the strong ability to translocate between cells[18]. Proteins fused with the protein transduction domain (PTD), which lies in C terminal of VP22 (34 amino acids), also can translocate into the target cells[24-26]. In this report, HBsAg concentration decreased 63.89% by PTD-HBc fusion based (pHVP22/D) DN mutant, and DN mutant with upstream 34 amino acids of VP22 (pHVP22/U, without PTD) only decreased 38.88%, suggesting that DN mutant with PTD can strongly inhibit the activity of wild-type viral gene than that without PTD, and the transduction domain of VP22 can enhance the antiviral effect of DN mutant. By detecting HBV-DNA concentrations in the supernatant, similar results could be obtained.
DN mutant based on full-length of VP22 (pHVP22/F) decreased HBsAg concentration and HBV-DNA content by 81.94% and 72.30%, respectively (Figures 3A and B), showing a significant difference compared to PTD-based DN mutant (pHVP22/D, P<0.05). Due to the different efficiency between the two constructs, the molecular mass of the protein fused to HBc C-terminal is related to the inhibitory effect of DN mutant. The larger the fused molecule, the stronger the antiviral effect of DN mutant. Possible explanations may be related to the stabilization of the larger fusion protein and the steric hindrance is necessary to inhibit proper assembly of the nucleocapsid.
In conclusion, DN mutant-based c-myc epitope (pHVP22/M) cannot inhibit HBV replication. Since c-myc epitope has only 10 amino acids, DN mutant of HBc may mediate antiviral effect through a minimal length of C-terminal. The related experiments are in process.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Fleming J. Current treatments for hepatitis. J Infus Nurs. 2002;25:379-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | elSaadany S, Tepper M, Mao Y, Semenciw R, Giulivi A. An epidemiologic study of hepatocellular carcinoma in Canada. Can J Public Health. 2002;93:443-446. [PubMed] |
| 3. | Mazumdar TN. Management of chronic hepatitis B infection: an update. J Indian Med Assoc. 2001;99:306-308, 310. [PubMed] |
| 4. | Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 827] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 5. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] |
| 6. | Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425-1430. [PubMed] |
| 7. | Manns MP. Current state of interferon therapy in the treatment of chronic hepatitis B. Semin Liver Dis. 2002;22 Suppl 1:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Shindo M, Hamada K, Koya S, Sokawa Y, Okuno T. The clinical significance of core promoter and precore mutations during the natural course and interferon therapy in patients with chronic hepatitis B. Am J Gastroenterol. 1999;94:2237-2245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Liaw YF. Therapy of chronic hepatitis B: current challenges and opportunities. J Viral Hepat. 2002;9:393-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Wolters LM, Hansen BE, Niesters HG, de Man RA. Viral dynamics in chronic hepatitis B patients treated with lamivudine, lamivudine-famciclovir or lamivudine-ganciclovir. Eur J Gastroenterol Hepatol. 2002;14:1007-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Santantonio T, Mazzola M, Iacovazzi T, Miglietta A, Guastadisegni A, Pastore G. Long-term follow-up of patients with anti-HBe/HBV DNA-positive chronic hepatitis B treated for 12 months with lamivudine. J Hepatol. 2000;32:300-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 185] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 12. | Chiou HC, Lucas MA, Coffin CC, Banaszczyk MG, Ill CR, Lollo CP. Gene therapy strategies for the treatment of chronic viral hepatitis. Expert Opin Biol Ther. 2001;1:629-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | von Weizsäcker F, Köck J, Wieland S, Offensperger WB, Blum HE. Dominant negative mutants of the duck hepatitis B virus core protein interfere with RNA pregenome packaging and viral DNA synthesis. Hepatology. 1999;30:308-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | von Weizsäcker F, Wieland S, Köck J, Offensperger WB, Offensperger S, Moradpour D, Blum HE. Gene therapy for chronic viral hepatitis: ribozymes, antisense oligonucleotides, and dominant negative mutants. Hepatology. 1997;26:251-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Scaglioni P, Melegari M, Takahashi M, Chowdhury JR, Wands J. Use of dominant negative mutants of the hepadnaviral core protein as antiviral agents. Hepatology. 1996;24:1010-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | von Weizsäcker F, Wieland S, Blum HE. Inhibition of viral replication by genetically engineered mutants of the duck hepatitis B virus core protein. Hepatology. 1996;24:294-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Scaglioni PP, Melegari M, Wands JR. Characterization of hepatitis B virus core mutants that inhibit viral replication. Virology. 1994;205:112-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 754] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 19. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A. 1987;84:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 939] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 20. | Acs G, Sells MA, Purcell RH, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci U S A. 1987;84:4641-4644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 120] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836-2844. [PubMed] |
| 22. | Trono D, Feinberg MB, Baltimore D. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell. 1989;59:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 231] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Malim MH, Freimuth WW, Liu J, Boyle TJ, Lyerly HK, Cullen BR, Nabel GJ. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197-1201. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 147] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Elliott G, O'Hare P. Intercellular trafficking of VP22-GFP fusion proteins. Gene Ther. 1999;6:149-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Aints A, Güven H, Gahrton G, Smith CI, Dilber MS. Mapping of herpes simplex virus-1 VP22 functional domains for inter- and subcellular protein targeting. Gene Ther. 2001;8:1051-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21:45-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 346] [Article Influence: 13.8] [Reference Citation Analysis (0)] |