Published online Jan 28, 2005. doi: 10.3748/wjg.v11.i4.503
Revised: March 21, 2004
Accepted: March 29, 2004
Published online: January 28, 2005
AIM: To explore the possibility of expressing hepatitis C virus (HCV) envelope protein 1 (E1) in Escherichia coli (E. coli) and to test the purified recombinant E1 proteins for clinical and research applications.
METHODS: C-terminally truncated E1 fragments were expressed in E. coli as hexa-histidine-tagged fusion proteins. The expression products were purified under denaturing conditions using immobilized-metal affinity chromatography. Purified E1 proteins were used to immunize rabbits. Rabbit anti-sera thus obtained were reacted with both E. coli- and mammalian cell-expressed E1 glycoproteins as detected by Western blot.
RESULTS: Full-length E1 protein proved difficult to express in E. coli. C-terminally truncated E1 was successfully expressed in E. coli as hexa-histidine-tagged recombinant fusion protein and was purified under denaturing conditions on Ni2+-NTA agarose. Rabbit anti-sera raised against purified recombinant E1 specifically reacted with mammalian cell-expressed E1 glycoproteins in Western blot. Furthermore, E. coli-derived E1 protein was able to detect animal antibodies elicited by E1-based DNA immunization.
CONCLUSION: These results demonstrate that the prokaryotically expressed E1 proteins share identical epitopes with eukaryotically expressed E1 glycoprotein. The E. coli-derived E1 proteins and corresponding antisera can become useful tools in anti-HCV vaccine research.
-
Citation: Liu J, Zhu LX, Kong YY, Li GD, Wang Y. Purification and application of C-terminally truncated hepatitis C virus E1 proteins expressed in
Escherichia coli . World J Gastroenterol 2005; 11(4): 503-507 - URL: https://www.wjgnet.com/1007-9327/full/v11/i4/503.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i4.503
Hepatitis C virus (HCV) is the major etiological agent of both community-acquired and post-transfusion non-A, non-B hepatitis[1,2]. It is estimated that 3% of world population have been infected with HCV[3]. Approximately 85% of patients develop chronic infection, and about 20% of chronic cases will progress into cirrhosis and/or hepatocellular carcinoma[4,5]. Presently, there is no vaccine against HCV[6] and the only available therapy, interferon-alpha on its own or in combination with ribavirin, is effective in only a minority of patients and carries the risk of serious side effects[7,8]. There is a pressing need to develop effective prophylactic and therapeutic measures against HCV in order to combat this global public health threat.
Envelope proteins of HCV (E1 and E2) are predicted to be type I membrane glycoproteins, and generally believed to constitute the protein components of virion membrane[9,10]. Various studies have implicated both E1 and E2 in important steps of HCV entry into target cells, such as receptor binding and membrane fusion[11-13]. Vaccination of chimpanzees with E1 and E2 glycoproteins resulted in limited but measurable protection against homologous virus challenge[14]. Therefore, E1 and E2 have become two major targets in HCV vaccine research.
Evaluation of HCV envelope protein-based vaccines requires an effective method for antigen detection and an inexpensive supply of large quantities of antigens. Although E1 glycoproteins expressed in mammalian cell systems in theory would best reflect properties of E1 proteins present on HCV virion membrane, low yield as well as difficulty in purification and scaling up makes such systems unsuitable for large-scale applications[14]. In order to circumvent this problem, we attempted to express E1 proteins in E. coli. Bacterial expression systems, compared to other higher organism expression systems, usually offer higher yield at considerably lower cost. Our previous work has shown that E. coli-derived recombinant E2 proteins and rabbit antisera against them are sufficient for these applications, and could partly substitute expensive mammalian system-expressed envelope proteins and infectious HCV patients’ sera[15-19]. In this work, C-terminally truncated E1 was expressed in E. coli as hexa-histidine-tagged fusion proteins and purified under denaturing conditions using immobilized-metal affinity chromatography. Rabbit anti-sera against E. coli-derived E1 proteins specifically reacted with mammalian cell-expressed E1 glycoproteins in Western blot. Furthermore, E. coli-derived E1 protein was able to detect animal antibodies elicited by E1-based DNA immunization. These results demonstrate that these prokaryotically expressed E1 proteins share similar epitopes with eukaryotically expressed E1 glycoproteins. The E. coli-derived E1 proteins and corresponding antisera could become useful tools in anti-HCV vaccine research.
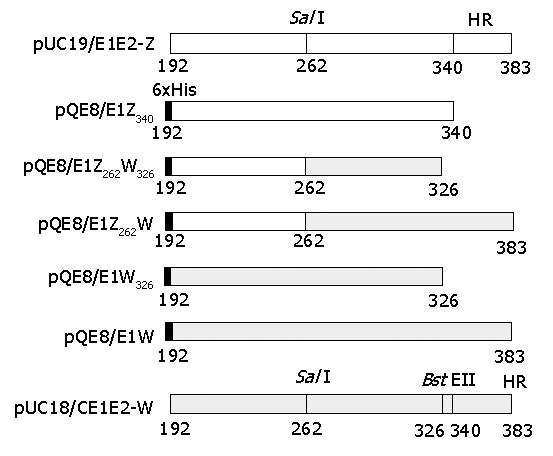
pUC18/CE1E2-W carrying C, E1 and E2 coding sequences of HCV (subtype 1b) was provided by professor Yu Wang of Peking University, Beijing (GenBank accession No. D10934)[20]. pUC19/E1E2-Z carrying E1 and E2 coding sequences of a different HCV isolate (also subtype 1b) was provided by Dr. Xin-Xin Zhang of Ruijin Hospital, Shanghai. pQE8 is an N-terminal hexa-histidine fusion expression vector from Qiagen GmbH, Hilden, Germany. E. coli strain TG-1 was used as cloning and expression host.
Polymerase chain reactions and recombinant cloning were performed according to standard protocols[21]. DNA sequences coding for E1 amino a-cid (aa) 192-340 and aa 192-383 were amplified from pUC19/E1E2-W and pUC18/CE1E2-Z, and cloned into pQE8 between the BamHI and HindIII sites to create pQE8/E1Z340 and pQE8/E1W respectively. Sequences between BamHI and SalI were removed from pQE8/E1Z340 by double digestion and used to replace corresponding sequences in pQE8/E1W. The resultant plasmid carrying chimeric E1 coding sequences was designated as pQE8/E1Z262W. Deletion of E1 C-terminal hydrophobic region (aa 341-383) in pQE8/E1W or pQE8/E1Z326W was conducted by double digestion with BstEII and HindIII, blunting of the resultant ends, removal of unwanted small fragments and self-ligation of large fragments. The obtained subclones were designated as pQE8/E1W326 and pQE8/E1Z262W326, respectively. Structures of these plasmids are illustrated in Figure 1.
Freshly saturated recombinant TG-1 culture was inoculated into fresh LB media at 1:100. Two hours after inoculation, expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mmol/L. Cells were harvested 6 h later by centrifugation and stored at -20 °C.
Solubility analysis and purification of expression products were performed as previously described[15-17]. Briefly, harvested bacteria were resuspended in phosphate-buffered saline (PBS, containing 137 mmol/L NaCl, 2.7 mmol/L KCl, 4.3 mmol/L Na2HPO4, 1.4 mmol/L KH2PO4, pH 7.3), sonicated on ice-bath, and centrifuged at 15000 r/min at 4 °C. The soluble and insoluble fractions after centrifugation were analyzed for the presence of expression products. Insoluble recombinant E1 proteins were extracted with 6 mol/L Gu•HCl/20 mmol/L β-ME/PBS (pH 8.0), centrifuged at 15000 r/min at 4 °C, and loaded onto pre-equilibrated Ni2+-NTA agarose (Qiagen). The gel matrices were sequentially washed with 6 mol/L Gu•HCl/20 mmol/L β-ME/PBS (pH 6.3) and 8 mol/L urea, 20 mmol/L β-ME/PBS (pH 6.3), and then eluted with 8 mol/L urea, 20 mmol/L β-ME/PBS (pH 4.3) or boiled in reductive SDS-PAGE sample buffer for elution.
Expression of HCV structural proteins C, E1 and E2 was performed as previously described[22]. Recombinant vaccinia virus vCEH-2 contained coding sequences of HCV polyprotein aa 1-730 under the control of T7 promoter, whereas vTT7 encoded the T7 polymerase required for expression. HeLa cells were co-infected with vTT7 and vCEH-2 at a multiplicity of infection of 4:4:1 (vTT7:vCEH-2:cell) and cultured for 48 h. Cells were collected by scraping, washed with PBS at 4 °C and stored at -20 °C.
SDS-PAGE under reducing or non-reducing conditions and Western blot were conducted according to standard protocols[22]. In Western blot, first antibody was diluted at 1:500 and second antibody [HRP-labeled protein A (Sigma, St. Louis, MO, USA) or swine anti-rabbit Ig (Dako, Denmark)] was diluted at 1:1000. Blots were developed using the enhanced chemi-luminescent (ECL) method (PerfectBio, Shanghai, China).
For deglycosylation analysis, cell samples were treated with PNGase F (New England Biolabs Inc., Beverly, MA, USA) according to manufacturer’s instructions and then subjected to reductive SDS-PAGE/Western blot analysis.
Female rabbits (Shanghai Laboratory Animal Center) were immunized subcutaneously on the back with 300 μg of purified recombinant protein emulsified in complete Freud’s adjuvant (Sigma) and boosted twice at an interval of 4 wk with the same quantity of antigen emulsified in incomplete Freud’s adjuvant (Sigma). One week after final boosting, total blood was collected through the carotid artery and serum was prepared using standard procedure[21].
Anti-E1 antibodies in post-immune animal sera were analyzed in standard ELISA using purified recombinant E1 protein as coating antigen. Microplates were coated with the antigen used for immunization at 0.1 μg/hole. Serially diluted post-immune sera were analyzed as previously described[15-17] with pre-immune sera diluted at 1:100 as negative control. The highest dilution giving a positive reading was taken as the antibody titer of corresponding antisera. All tests were done in duplicate.
For the expression of HCV E1 protein in E. coli, full-length or C-terminally truncated E1 sequences from two subtype 1b isolates were used (Figure 1). Chimeric constructs carrying sequences derived from both isolates were also used (Figure 1). Corresponding coding sequences were cloned into N-terminal hexa-histidine fusion expression vector pQE8 as described in MATERIALS AND METHODS.
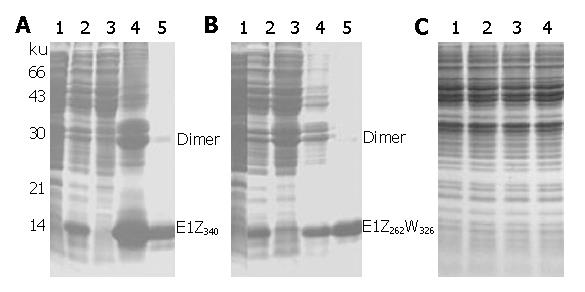
TG-1 cells transformed with recombinant expression plasmids were induced with IPTG and analyzed on SDS-PAGE (Figure 2). C-terminally truncated pUC19/E1E2-Z-derived pQE8/E1Z340 expressed high levels of recombinant E1 protein of predicted apparent molecular weight (Figure 2A, lane 2), whereas pUC18/CE1E2-W-derived full-length pQE8/E1W or C-terminally truncated pQE8/E1W326 displayed no obvious expression of recombinant proteins (Figure 2C). However, when N-terminal sequences (aa 192-262) in pQE8/E1W326 were replaced by corresponding pUC19/E1E2-Z-derived sequences, the resultant chimeric pQE8/E1Z262W326 showed prominent expression of recombinant E1 proteins upon induction (Figure 2B, lane 2). Expression products from pQE8/E1Z340 and pQE8/E1Z262W326 were designated as E1Z340 and E1Z262W326, respectively.
Solubility analysis revealed that both E1Z340 and E1Z262W326 were highly insoluble (Figures 2A and 2B, lanes 3 and 4) and required high concentration of strong chaotropic agent (6 mol/L Gu•HCl) for efficient extraction (data not shown). Solubilized E1Z340 and E1Z262W326 were purified on Ni2+-NTA agarose under denaturing conditions. Purified E1Z340 was eluted using low pH, whereas purified E1Z262W326 was eluted either using low pH or by boiling the gel matrices in reductive SDS-PAGE sample buffer, and designated as E1Z262W326N and E1Z262W326R respectively. Two types of E1Z262W326 eluted using different methods displayed similar electrophoresis patterns in SDS-PAGE (representative data obtained with E1Z262W326N are shown in Figure 2B, lanes 4 and 5) and almost identical reactivity patterns in Western blot (see below). After Coomassie brilliant blue staining, both purified E1Z340 and E1Z262W326 displayed minor quantities of higher and lower mobility bands, in addition to the major full-length monomer bands of 18 ku and 16.8 ku respectively (Figure 2A, lane 5; and Figure 2B, lane 5). These might correspond to dimers of the monomer form and non-full-length forms with incomplete C-termini, possibly a result of premature translational termination or enzyme degradation. The purity of finally obtained E1Z340 and E1Z262W326 exceeded 85% by densitometric scanning and the yield of both proteins was estimated to be above 1 mg/L LB culture.
Purified E1Z262W326 was used to detect anti-E1 antibodies in E1-based DNA immunization studies. Sera from BALB/c mice injected with E1-expressing plasmids specifically reacted with E1Z262W326 in enzyme-linked immunosorbent assay (ELISA), as previously reported.
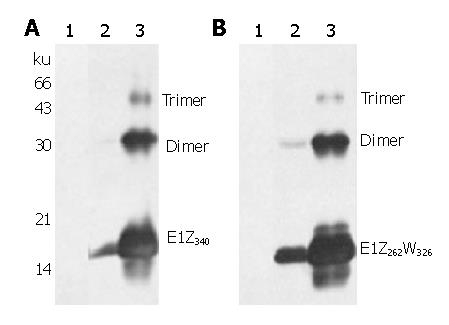
Purified E1Z340, E1Z262W326N and E1Z262W326R were used to immunize rabbits and the obtained antisera were designated as RE1Z340, RE1Z262W326N and RE1Z262W326R respectively. Their anti-E1 titers were determined by ELISA to be 1: 1×103, 1: 8×104 and 1: 1.6×105 respectively.
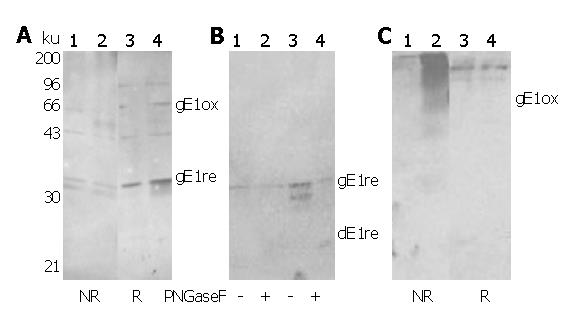
All three rabbit anti-E1 sera displayed similar recognition of E. coli-expressed E1 proteins with high sensitivity and specificity. Representative data obtained from RE1Z340 and RE1Z262W326N are shown in Figure 3. For glycosylated E1 proteins expressed using recombinant vaccinia virus system, RE1Z340 showed no specific recognition (data not shown), whereas RE1Z262W326R displayed specific recognition of both oxidized and reduced E1 glycoproteins (Figure 4A). RE1Z262W326N, in contrast, only recognized oxidized E1 glycoproteins with high sensitivity (Figure 4C). Both RE1Z262W326R and RE1Z262W326N recognized deglycosylated E1 glycoproteins, as shown representatively in Figure 4B.
Various reports have suggested that, in addition to being a structural component of HCV virion membrane, E1 also plays important roles in viral attachment and entry into target cells as well as virus-host immune interactions[11-14]. As a result, both virologists and vaccinologists are paying considerable attention to this protein.
It has been reported that the C-terminal hydrophobic region (aa 341-383) of E1 hinders its expression in E. coli, possibly through interference with normal bacterial membrane functions[23-25]. In our work, we also found that constructs carrying the hydrophobic region were unable to express recombinant proteins to a level detectable by Coomassie brilliant blue staining (Figure 2C). Truncation of the hydrophobic region resulted in expression of high levels of recombinant proteins upon induction, but only for those constructs carrying N-terminal sequences (aa 192-262) derived from pUC19/E1E2-Z, i.e., pQE8/E1Z340 and pQE8/E1Z262W326. The construct carrying pUC19/CE1E2-W-derived N-terminal sequences (pQE8/E1W326) showed no difference in its full-length predecessor pQE8/E1W (Figure 2). Comparison of pUC19/E1E2-Z and pUC19/CE1E2-W sequences in the aa 192-262 region revealed 14 transitions and 4 transversions, resulting in four amino acid residue changes (data not shown), suggesting that nucleotide and/or amino acid sequences play an important role in E1 expression in E. coli. A similar phenomenon was also observed when we attempted to express HCV E2 in E. coli[16]. The exact mechanism of sequence difference causing different expression is still under investigation.
Expression products induced from pQE8/E1Z340 and pQE8/E1Z262W326 are largely insoluble, which is in agreement with reports by other researchers[24,26]. Large-scale expression and purification using denaturing immobilized-metal affinity chromatography could produce large quantities (>1 mg/L) of both E1Z340 and E1Z262W326 with fairly high purity (>85%).
Rabbit sera raised against these proteins not only recognized E. coli-derived E1 proteins with extremely high specificity (Figure 3), but also are capable of detecting subtype 1b E1 glycoproteins expressed in mammalian cells (Figure 4 and results reported elsewhere[27]). These results suggest that bacterially-derived E1 proteins share identical epitopes with mammalian E1 glycoproteins, and that these epitopes can be presented to the host immune system in vivo. It has been reported that mutation of N-glycosylation sites on E1 sometimes improves its immunogenicity[28]. In light of this, although envelop-targeted HCV vaccine research has almost exclusively focused on glycosylated proteins expressed in mammalian cells, it would be interesting to test E. coli-derived non-glycosylated E1 proteins reported here as a possible HCV vaccine candidate.
Rabbit anti-E1Z340 antisera did not react with mammalian E1 glycoproteins as detected by Western blot, possibly as a result of its relatively low anti-E1 titer. Two types of reactivity pattern were observed for rabbit anti-E1Z262W326 antisera: antisera raised against E1Z262W326 eluted under reducing conditions (RE1Z262W326R) displayed specific recognition of both oxidized and reduced forms of E1 glycoproteins (Figure 4A), whereas antisera raised against E1Z262W326 eluted under non-reducing conditions (RE1Z262W326N) only recognized oxidized forms of E1 glycoproteins (Figure 4C). This result suggests that some disulfide bonds formed in E. coli or during extraction-purification procedures fold E1 polypeptide to present non-linear epitopes identical to those found on mammalian E1 glycoproteins. Such reactivity against non-reduced E1 glycoproteins makes these sera promising candidates for the histological detection of E1 in liver biopsy samples.
Since bacteria-derived E1 proteins share identical epitopes with mammalian E1 glycoproteins, we used E1Z262W326 to detect anti-E1 antibodies in E1-based DNA immunization studies. BALB/c mice immunized with E1-expressing plasmids developed anti-E1 antibodies detectable by E1Z262W326 in ELISA[24]. This result further demonstrates the immunogenic/antigenic similarity between bacterial and mammalian E1 proteins. We also tested purified E1 proteins for their ability to react with two homologous patients’ sera in Western blot or ELISA. With the two sera we used, no specific reaction could be obtained. The failure of E. coli-derived E1 proteins to react with homologous patients’ sera might indicate that these sera do not contain any anti-E1-polypeptide antibodies, or the antibody titer is too low to be detected. In chronic patients, it has been reported that the prevalence of anti-E1-polypeptide antibody is only about 51.5%[26] or even lower[29].
In summary, this work produced large quantities of E. coli-derived C-terminally truncated E1 proteins with high purity, obtained highly specific rabbit antisera against these proteins, and demonstrated the applicability of these sera for the detection of E1 proteins expressed in both prokaryotic and eukaryotic systems. These results indicate that E. coli-derived E1 proteins are immunologically similar to mammalian E1 glycoproteins. E1 antigens and antisera reported here can serve as useful tools in the development of E1-based HCV vaccine as well as other E1-related studies.
The authors are grateful to professor Yu Wang and Dr. Xin-Xin Zhang for providing the HCV cDNA plasmids and homologous patients’ sera.
Edited by Kumar M and Wang XL Proofread by Zhu LH
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4655] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 2. | Cohen J. The scientific challenge of hepatitis C. Science. 1999;285:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 273] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 3. | Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat. 1999;6:35-47. [PubMed] |
| 5. | Wawrzynowicz-Syczewska M, Kubicka J, Lewandowski Z, Boroń-Kaczmarska A, Radkowski M. Natural history of acute symptomatic hepatitis type C. Infection. 2004;32:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Kew M, François G, Lavanchy D, Margolis H, Van Damme P, Grob P, Hallauer J, Shouval D, Leroux-Roels G, Meheus A. Prevention of hepatitis C virus infection. J Viral Hepat. 2004;11:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [PubMed] |
| 8. | Zein CO, Zein NN. Advances in therapy for hepatitis C infection. Microbes Infect. 2002;4:1237-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 10. | Selby MJ, Choo QL, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Garcia JE, Puentes A, Súarez J, López R, Vera R, Rodríguez LE, Ocampo M, Curtidor H, Guzman F, Urquiza M. Hepatitis C virus (HCV) E1 and E2 protein regions that specifically bind to HepG2 cells. J Hepatol. 2002;36:254-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 12. | Lagging LM, Meyer K, Owens RJ, Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J Virol. 1998;72:3539-3546. [PubMed] |
| 13. | Triyatni M, Saunier B, Maruvada P, Davis AR, Ulianich L, Heller T, Patel A, Kohn LD, Liang TJ. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J Virol. 2002;76:9335-9344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 397] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 15. | Liu J, Zhu L, Zhang X, Lu M, Kong Y, Wang Y, Li G. Expression, purification, immunological characterization and application of Escherichia coli-derived hepatitis C virus E2 proteins. Biotechnol Appl Biochem. 2001;34:109-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Liu J, Kong Y, Zhu L, Wang Y, Li G. High-level expression of the C-terminal hydrophobic region of HCV E2 protein ectodomain in E. coli. Virus Genes. 2002;25:5-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 17. | Liu J, Zhang XX, Zhang SY, Lu M, Kong YY, Wang Y, Li GD. Expression of hepatitis C virus E2 ectodomain in E. coli and its application in the detection of anti-E2 antibodies in human sera. Acta Biochim Biophys Sin (Shanghai). 2004;36:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Zhu LX, Liu J, Li YC, Kong YY, Staib C, Sutter G, Wang Y, Li GD. Full-length core sequence dependent complex-type glycosylation of hepatitis C virus E2 glycoprotein. World J Gastroenterol. 2002;8:499-504. [PubMed] |
| 19. | Wang CL, Zhu LX, Liu J, Zhang ZC, Wang Y, Li GD. Expression and characterization of hepatitis C Virus E2 glycoprotein fused to hepatitis B virus preS1(21-47) fragment in CHO cells. Shengwuhuaxue Yu Shengwuwuli Xuebao (Shanghai). 2002;34:400-404. [PubMed] |
| 20. | Wang Y, Okamoto H, Tsuda F, Nagayama R, Tao QM, Mishiro S. Prevalence, genotypes, and an isolate (HC-C2) of hepatitis C virus in Chinese patients with liver disease. J Med Virol. 1993;40:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press 2001; . |
| 22. | Li YC, Li GC, Kong YY, Wang Y, Wang Y, Wen YM. Expression of structural proteins of hepatitis C virus (HCV) in mammalian cells. Zhongguo Kexue. 1998;28:204-210. |
| 23. | Yan BS, Liao LY, Leou K, Chang YC, Syu WJ. Truncating the putative membrane association region circumvents the difficulty of expressing hepatitis C virus protein E1 in Escherichia coli. J Virol Methods. 1994;49:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Ciccaglione AR, Marcantonio C, Costantino A, Equestre M, Geraci A, Rapicetta M. Expression and membrane association of hepatitis C virus envelope 1 protein. Virus Genes. 2000;21:223-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Montigny C, Penin F, Lethias C, Falson P. Overcoming the toxicity of membrane peptide expression in bacteria by upstream insertion of Asp-Pro sequence. Biochim Biophys Acta. 2004;1660:53-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Lorenzo LJ, García O, Acosta-Rivero N, Dueñas-Carrera S, Martínez G, Alvarez-Obregón J, Pichardo D, Ramos A, Guerra I, Morales J. Expression and immunological evaluation of the Escherichia coli-derived hepatitis C virus envelope E1 protein. Biotechnol Appl Biochem. 2000;32:137-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Zhu J, Kong YY, Liu J, Zhang ZC, Wang Y, Li GD. Secretory Expression of Different C-terminal Truncated HCV E1 Proteins in Mammalian Cells and Characterization of the Expressed Products. Shengwuhuaxue Yu Shengwuwuli Xuebao (Shanghai). 2001;33:634-640. [PubMed] |
| 28. | Fournillier A, Wychowski C, Boucreux D, Baumert TF, Meunier JC, Jacobs D, Muguet S, Depla E, Inchauspé G. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J Virol. 2001;75:12088-12097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Hada H, Koide N, Hanafusa T, Sakaguchi K, Shinji T, Sasaki S, Oka T, Takayama N, Yumoto Y, Tsuji T. Detection by western blotting of an antibody to the hepatitis C virus E1 envelope protein in sera of patients with chronic liver disease. Acta Med Okayama. 1992;46:365-370. [PubMed] |