Published online Oct 7, 2005. doi: 10.3748/wjg.v11.i37.5811
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: October 7, 2005
AIM: To establish the rapid, specific, and sensitive method for detecting O157:H7 with DNA microchips.
METHODS: Specific oligonucleotide probes (26-28 nt) of bacterial antigenic and virulent genes of E. coli O157:H7 and other related pathogen genes were pre-synthesized and immobilized on a solid support to make microchips. The four genes encoding O157 somatic antigen (rfbE), H7 flagellar antigen (fliC) and toxins (SLT1, SLT2) were monitored by multiplex PCR with four pairs of specific primers. Fluorescence-Cy3 labeled samples for hybridization were generated by PCR with Cy3-labeled single prime. Hybridization was performed for 60 min at 45 °C. Microchip images were taken using a confocal fluorescent scanner.
RESULTS: Twelve different bacterial strains were detected with various combinations of four virulent genes. All the O157:H7 strains yielded positive results by multiplex PCR. The size of the PCR products generated with these primers varied from 210 to 678 bp. All the rfbE/fliC/SLT1/SLT2 probes specifically recognized Cy3-labeled fluorescent samples from O157:H7 strains, or strains containing O157 and H7 genes. No cross hybridization of O157:H7 fluorescent samples occurred in other probes. Non-O157:H7 pathogens failed to yield any signal under comparable conditions. If the Cy3-labeled fluorescent product of O157 single PCR was diluted 50-fold, no signal was found in agarose gel electrophoresis, but a positive signal was found in microarray hybridization.
CONCLUSION: Microarray analysis of O157:H7 is a rapid, specific, and efficient method for identification and detection of bacterial pathogens.
-
Citation: Jin HY, Tao KH, Li YX, Li FQ, Li SQ. Microarray analysis of
Escherichia coli O157:H7. World J Gastroenterol 2005; 11(37): 5811-5815 - URL: https://www.wjgnet.com/1007-9327/full/v11/i37/5811.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i37.5811
In 1982, two outbreaks of hemorrhagic colitis occurred among patrons of a fast-food restaurant chain in Oregon and Michigan. The disease is characterized by watery diarrhea progressing rapidly to grossly bloody diarrhea and abdominal cramping with no fever. An investigation by state health departments and Centers for Disease Control (CDC) indicates that E. coli O157:H7, a rare strain not previously linked to human disease, is the cause of the disease[1]. In 1983, Karmali et al[2] reported that E. coli O157:H7 is related with hemolytic uremic syndrome (HUS), a leading cause of kidney failure in children. Since then, E. coliO157:H7 has been recognized as the leading cause of HUS in the USA, Japan, and in our country. Infection with E. coliO157:H7 can be entirely asymptomatic or presents with watery diarrhea, bloody diarrhea, HUS, thrombotic thrombocytopenic purpura, leading to death[3]. Antimicrobial agents have no proven value in the treatment of E. coli O157:H7 infections. It was reported that patients receiving antimicrobial agents are more likely to develop HUS than patients not receiving such drugs[4]. It may be prudent to avoid the use of antimicrobial agents in patients with diagnosed or suspected E. coli O157:H7 infection. Traditionally, E. coli O157:H7 is identified by microbiological culture techniques and immunological methods that are used to detect the O157 and H7 antigens[5]. These methods are slow and complicated, and sometimes yield false-positive results when other variants of E. coli O157:H7 are occasionally isolated in the initial culture. Therefore, accurate and rapid identi-fication is important for the diagnosis of O157:H7 infection.
DNA and oligonucleotide microchip (microarray) technology play an increasingly important role in gene expression, phylogenetic classification of bacteria, mapping of genes, analysis of single-nucleotide polymorphisms, drug discovery, multiple microbial diagnosis, and toxicological research[6-8]. Since microchips with short (8-30 nt) synthetic oligonuc-leotides are more sensitive to genetic differences, they are more appropriate for discriminating microorganisms.
In the present study, we used gene rfbE encoding an enzyme involved in the biosynthesis of O157 antigen (the somatic antigen), and gene fliC encoding the H7 flagellar antigen, and gene Shiga-like toxin (SLT) 1 and 2, encoding virulent factors, for the diagnosis of E. coli O157:H7 infection diagnosis by microarray analysis. It seems very useful for automatic identification and characterization of bacterial pathogens.
O157:H7, Vibrio cholerae, Shigella, Salmonella, DH5a, and other E. coli strains were from National Institute for the Control of Pharmaceutical and Biological Products, CDC of Jiangsu and Shandong Provinces, and our laboratory. All strains were stored at -80 °C in Luria-Bertani broth containing 15% glycerol. Schistosoma japonicum, hemorrhagic fever and renal syndrome virus (HFRSV), Leptospira interrogans, Rickettsia Tsutsugamushi, and Plasmodium were also from CDC of Jiangsu, Guangdong, Fujian Provinces and our laboratory.
Primers used to amplify the fragments of antigenic genes and virulent genes are shown in Table 1. One primer was labeled with Cy3 (TaKaRa Co.). BLAST search was used to retrieve the homologous sequences of each of the four genes analyzed. For each gene, one conserved region containing approximately 26-28 nucleotides was chosen as the target sequence probe. The probes and their BLAST search results are also shown in Table 1.
| Targetgene | Primer sequences forPCR amplification | Labeledprimer | PCR productsize (bp) | Oligonucleotides targetadsorbed by array chip | GenBank No. ofBLAST search |
| O157 | L5’AACGGTTGCTCTTCATTTAG3’ | R | 678 | GGTGGAATGGTTGTCACGAATGACAAA | AE005429, AP002559, AB008676 |
| (rfbE) | R5’GAGACCATCCAATAAGTGTG3’ | AF061251 S83460 | |||
| H7 | L5’TACCACCAAATCTACTGCTG3’ | R | 560 | GACGATGCAGGCAACTTGACGACTAA | AE005415, AF228496, U47614 |
| (fliC) | R5’TACCACCTTTATCATCCACA3’ | AP002559, AB028474, L07388 | |||
| SLT1 | L5’TGTAACTGGAAAGGTGGAGTATAC3’ | R | 210 | GGATGACGTAACCATTAAAACTAATGCC | AY123842, AF461172,AJ251754, |
| R5’GCTATTCTGAGTCAACGAAAAATAAC3’ | AB071624, AE005442,X07903 | ||||
| SLT2 | L5’TTTACGATAGACTTCTCGAC3’ | L | 228 | AGATGGTCAAAACGCGCCTGATAGAC | AF329817, AE005296, AJ272135 |
| R5’CACATATAAATTATTTCGCTC3’ | AB052227, AP002554, Y10775 |
Microchips were printed on silylated (aldehyde-coated) glass slides (8 cm2.5 cm, Sigma Co.) by the contact microspotting robotic system 7 500, the PixSys 5 500 workstation microspotting software (Cartesian Technologies, Inc.), and the ChipMaker Pin 40 microspotting device. Microchip design maps are shown in Table 2. The final spotting mixture contained 100 mmol/L specific oligonucleotide probe (TaKaRa Co.) in 0.25 mol/L acetic acid. A range of 70-75% humidity was maintained in an environmental chamber to prevent rapid evaporation of spotted drops and efficient coupling of amino-modified oligonucleotides with the aldehyde groups on the glass surface. Six identical probe area arrays were printed on each slide, allowing analysis of six samples on one glass slide simultaneously. The size of spots did not exceed 150 mm in diameter. Printed slides were incubated at 37 °C for 60 min, then overnight at room temperature. The next day, slides were washed twice with 0.2% sodium dodecyl sulfate (SDS) in water for 5 min, twice with distilled water for 5 min each, and then dried in air. Before hybridization, to make the bonds between oligonucleotides and glass surface irreversible, the slides were incubated for 30 min in a freshly prepared 0.6% aqueous solution of NaBH4, then washed twice with 0.2% SDS for 1 min and twice with distilled water for 1 min. Microchips prepared according to this protocol could be stored for 2 mo at room temperature.
| 16 3×SSC | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 8 Vibrio cholerae ompW |
| 15 3×SSC | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 7 Schistosoma japonicum |
| 14 random probe | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 6 Plasmodium V |
| 13 SLT1 gene | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 5 Plasmodium F |
| 12 SLT2 gene | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 4 Leptospira interrogans |
| 11 fliC gene (H7) | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 3 Rickettsia tsutsugamushi |
| 10 rfbE gene (O157) | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 2 HFRSV 1 |
| 9 Vibrio cholerae ctx | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | 1 HFRSV 2 |
E. coli cultures were grown overnight at 37 °C in Luria-Bertani broth. One milliliter of the culture sample was centrifuged, and the pellet was washed and resuspended in 100 mL of distilled water. The suspension was boiled for 10 min followed by centrifugation at 10 000 g for 10 min to remove denatured proteins and bacterial cell membranes. Five-microliter aliquot of the supernatant was used as the template for multiplex PCR amplification. Briefly, 50 mL of the reaction mixture containing 5 mL,10 × PCR buffer, 0.2 mmol/L dNTPs, 2.0 mmol/L MgCl2, 0.5-1.0 pmol/L each primer and 1.5 U of Taq polymerase (TaKaRa Co.) was used. Temperature conditions consisted of an initial pre-denaturation step at 95 °C for 5 min, followed by 35 cycles at 94°C for 30 s, at 57 °C for 45 s, and at 72 °C for 60 s, a final extension at 72 °C for 5 min. The PCR products were analyzed by electrophoresis on 1% agarose gel in TAE (40 mmol/L Tris-acetate, 2 mmol/L disodium EDTA). DNA bands in the gels were stained with 0.5 mg/mL of ethidium bromide. The interested DNAs were extracted using DNA fragment purification kit Ver.2.0 (TaKaRa Co.) and dissolved in 50 mL of distilled water for the next Cy3 labeling.
Fluorescence-labeled samples for hybridization were generated by PCR as described above, except that the annealing temperature was 42 °C, and the template was the first multiplex PCR products (5 mL), the primer was only one Cy3-labeled (0.2 mmol/L) primer or four different Cy3-labeled (0.2-0.4 mmol/L) primers.
Immediately before hybridization, 8 mL Cy3-labeled PCR samples was mixed with 3 mL hybridization buffer containing Cy3-labeled quality control (0.3 mmol/L) probe, and denatured for 5 min at 95 °C, then chilled on ice for 1 min. This conc-entration could ensure rapid and efficient hybridization of a fluorescent sample with immobilized probes. Five-microliter aliquots from each sample was applied to the microarray area and covered with a glass coverslip to prevent evaporation of the sample during hybridization in the incubation chamber. Hybridization was performed for 60 min at 45 °C. Slides were then washed twice with 2 × SSC containing 0.1% SDS for 5 min at 60 °C, twice with 0.1 × SSC containing 0.1% SDS for 5 min at room temperature, and once with 0.1 × SSC, once with distilled water and dried by an air stream to completely remove any remaining solution. Then microchip images were taken with the confocal fluorescent scanner ScanArray 3000.
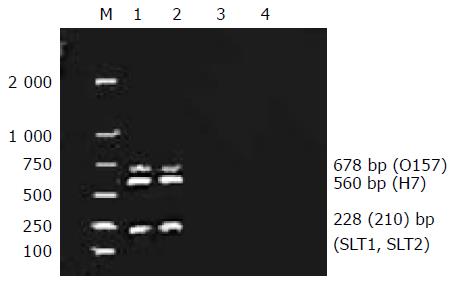
We detected 12 different bacterial strains with various combinations of four virulent genes. The results of multiplex PCR and microchip analysis generated by bacterial DNA and four primers or probes are shown in Table 3. All the O157:H7 strains yielded positive results. The size of the PCR products generated by these primers varied from 210 to 678 bp. The other non-O157:H7 strains were not positive. The pitfalls of agarose gel analysis of multiplex PCR are illustrated in Figure 1. The difference of SLT1 and SLT2 was too small (only 18 bp) to be distinguished in agarose gel electrophoresis.
| Strain | PCR | Chipping | ||||||
| rfbE | fliC | SLT1 | SLT2 | rfbE | fliC | SLT1 | SLT2 | |
| O157:H7 933 | + | + | + | + | + | + | + | |
| O157:H7 882364 | + | + | + | + | + | + | + | |
| O157:H19 44752 | + | - | - | - | + | - | - | - |
| O1:H7 44766 | - | + | - | - | - | + | - | - |
| O119:H6 44146 | - | - | - | - | - | - | - | - |
| Vibrio cholerae16017 | - | - | - | - | - | - | - | - |
| Shigella dysenteriae51252 | - | - | - | - | - | - | - | - |
| Shigella flexneri51302 | - | - | - | - | - | - | - | - |
| Shigella boydii51315 | - | -- | - | - | - | - | - | - |
| Shigella sonnei51334 | - | - | - | - | - | - | - | - |
| Salmonella 50087 | - | - | - | - | - | - | - | - |
| DH5a | - | - | - | - | - | - | - | - |
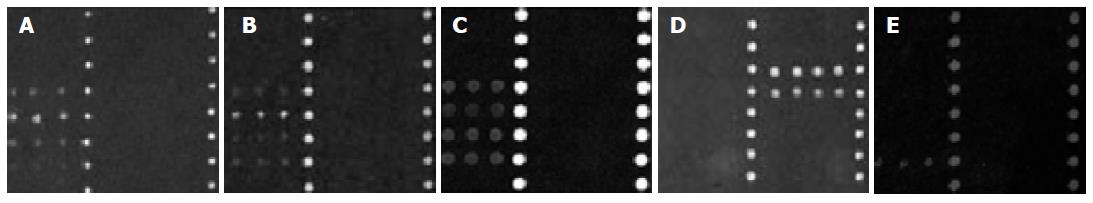
All the rfbE/fliC/SLT1/SLT2 probes specifically recognized Cy3-labeled fluorescent samples from O157:H7 species, or strains containing O157 and H7 genes. Neither the cross hybridization of O157:H7 fluorescent samples to other probes (Figures 2A-C), nor cross hybridization of rfbE/fliC/SLT1/SLT2 probes to other pathogens (Figure 2D) was detected. If the Cy3-labeled fluorescent product of O157 single PCR was diluted 50-fold, no signal was visible in agarose gel electrophoresis, but a positive signal was found in microarray hybridization (Figure 2E).
E. coli is a normal inhabitant in the intestinal tract of human beings and warm-blooded animals. Although usually harmless, various strains such as enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC), have virulent genes and render them pathogenic for both human beings and animals. E. coli O157:H7 is an important cause of hemorrhagic colitis and HUS. Conventional methods for detecting O157:H7 involve growth of non-sorbitol-fermenting E. colicolonies on sorbitol MacConkey agar culture (SMAC)[9]. These methods are time consuming, laborious, and expensive. More importantly, routine biochemical and serologic tests for pathogen identification can only type the species or serogroup level, but cannot directly characterize virulent genes. Thus, these assays cannot provide any information about the potential pathogenicity or virulence of O157:H7[10]. Several genetic markers such as rfbE[11], fliC[12], and certain virulent genes as attachment and effacing gene (eaeA), hemolysin A (hlyA)[13,14], SLT1, SLT2[15] are useful targets for multiplex PCR amplification and differentiation of O157:H7. Although multiplex PCR amplification is a simple and sensitive method[16], it has some inherent shortcomings. When multiplex PCR is used for detection of several genetic markers, it tends to generate more nonspecific DNA products. If the size of PCR products is similar, it is difficult to discriminate these products. Therefore, it is necessary to establish high-throughput specific and sensitive methods for detecting O157:H7.
Hybridization of DNA samples to miniature arrays (microchips) of immobilized gene-specific DNA or oligonuc-leotide probes has recently become a powerful tool. A number of different ways can make oligonucleotide microchips, including in situsynthesis of oligonucleotides directly on the microchip surface[17] and immobilization of pre-synthesized oligonucleotides on a solid support[18]. We designed four genotype-specific oligonucleotide probes (rfbE, fliC, SLT1, and SLT2) for O157:H7, and immobilized them on a modified glass surface to create the microchip. RfbE and fliC are the specific marker genes encoding O157 somatic antigen and H7 flagellar antigen. SLT1 and SLT2 are the important virulent genes. Schmidt et al[19] reported that the bacteriophage-encoded SLT plays a very important role in the pathogenesis of diseases caused by O157:H7, and in HUS through its cytotoxic effects on cells of the kidneys, intestines, central nervous system, and other organs. If infected with O157:H7 strain with SLT, many patients would develop HUS. Using these microchips, we can not only characterize the species of pathogens, but also identify their virulent factors SLT1 and SLT2. The major advantage of the microarray assay over agarose gel analysis of multiplex PCR products is that the microarray requires the internal sequences of DNA fragments to be complementary to the oligonucleotide probes on the microchip, but does not solely rely on the length of PCR products. In our study, only O157:H7 strains and SLT1, SLT 2 presented positive fluorescent signals. The successful demonstration of hybridization specificity led us to address the second critical parameter of microarray, assay sensitivity. When the Cy3-labeled product was diluted 50-fold, no positive signal was visible in agarose gel electrophoresis, but a positive hybridization signal was found in microchip, suggesting that microarray is more sensitive than agarose gel analysis.
In this study, to assess the synthesis of Cy3-labeled samples by multiplex and single PCR, we examined the sameO157:H7 882364 strain in different ways. Multiplex and single PCR could display positive hybridization in microchip. Compared to multiplex PCR, the fluorescent signals of single PCR synthesis were more homogeneous and easier to control the PCR mixture concentration and microarray sample content. In conclusion, microarray analysis is a fast, sensitive, specific, and high throughput method for detecting O157:H7.
Science Editor Wang XL and Li WZ Language Editor Elsevier HK
| 1. | Besser RE, Griffin PM, Slutsker L. Escherichia coli O157: H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu Rev Med. 1999;50:355-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 130] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Karmali MA, Steele BT, Petric M, Lim C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;1:619-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 644] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 3. | Ibekwe AM, Watt PM, Grieve CM, Sharma VK, Lyons SR. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157: H7 in dairy wastewater wetlands. Appl Environ Microbiol. 2002;68:4853-4862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 170] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 4. | Guidance for the diagnosis and management of suspected or proven Escherichia coli O157 infection. Prepared by the Scottish Infection Standards and Strategy Group and supported by the Scottish Executive Health Department. J R Coll Physician Edinb. 2004;34:37-40. |
| 5. | Nei QH. Infection of Escherichia coli O157: H7. Shijie Huaren Xiaohua Zazhi. 2001;9:944-945. |
| 6. | Hinchliffe SJ, Isherwood KE, Stabler RA, Prentice MB, Rakin A, Nichols RA, Oyston PC, Hinds J, Titball RW, Wren BW. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 2003;13:2018-2029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 128] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 7. | McCaffrey RL, Fawcett P, O'Riordan M, Lee KD, Havell EA, Brown PO, Portnoy DA. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386-11391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 140] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 8. | Shioda T. Application of DNA microarray to toxicological research. J Environ Pathol Toxicol Oncol. 2004;23:13-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Cody SH, Glynn MK, Farrar JA, Cairns KL, Griffin PM, Kobayashi J, Fyfe M, Hoffman R, King AS, Lewis JH. An outbreak of Escherichia coli O157: H7 infection from unpasteurized commercial apple juice. Ann Intern Med. 1999;130:202-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 244] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Holland JL, Louie L, Simor AE, Louie M. PCR detection of Escherichia coli O157: H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J Clin Microbiol. 2000;38:4108-4113. [PubMed] |
| 11. | Fortin NY, Mulchandani A, Chen W. Use of real-time polymerase chain reaction and molecular beacons for the detection of Escherichia coli O157: H7. Anal Biochem. 2001;289:281-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Wang L, Rothemund D, Curd H, Reeves PR. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157: H7. J Clin Microbiol. 2000;38:1786-1790. [PubMed] |
| 13. | Blanco M, Blanco JE, Mora A, Rey J, Alonso JM, Hermoso M, Hermoso J, Alonso MP, Dahbi G, González EA. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J Clin Microbiol. 2003;41:1351-1356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 159] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 14. | Fagan PK, Hornitzky MA, Bettelheim KA, Djordjevic SP. Detection of shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65:868-872. [PubMed] |
| 15. | Call DR, Brockman FJ, Chandler DP. Detecting and genotyping Escherichia coli O157: H7 using multiplexed PCR and nucleic acid microarrays. Int J Food Microbiol. 2001;67:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 103] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Syrmis MW, Whiley DM, Thomas M, Mackay IM, Williamson J, Siebert DJ, Nissen MD, Sloots TP. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004;6:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 17. | Dolan PL, Wu Y, Ista LK, Metzenberg RL, Nelson MA, Lopez GP. Robust and efficient synthetic method for forming DNA microarrays. Nucleic Acids Res. 2001;29:E107-E107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Okamoto T, Suzuki T, Yamamoto N. Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat Biotechnol. 2000;18:438-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Schmidt H, Scheef J, Huppertz HI, Frosch M, Karch H. Escherichia coli O157: H7 and O157: H(-) strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3491-3496. [PubMed] |