Published online Aug 14, 2005. doi: 10.3748/wjg.v11.i30.4693
Revised: December 15, 2004
Accepted: December 20, 2004
Published online: August 14, 2005
AIM: To elucidate the relationship between the frequency of core mutations and the clinical activity of hepatitis B virus (HBV)-related liver disease and to characterize the amino acid changes in the core region of HBV.
METHODS: We studied 17 Chinese patients with chronic hepatitis B according to their clinical courses and patterns of the entire core region of HBV.
RESULTS: Amino acid changes often appeared in the HBV core region of the HBV gene in patients with high values of alanine aminotransferase (ALT) or with the seroconversion from HbeAg to anti-HBe. The HBV core region with amino acid changes had high frequency sites that corresponded to HLA I/II restricted recognition epitopes reported by some investigators.
CONCLUSION: The core amino acid changes of this study occur due to influence of host immune system. The presence of mutations in the HBV core region seems to be important for predicting the clinical activity of hepatitis B in Chinese patients.
- Citation: Tanaka H, Ueda H, Hamagami H, Yukawa S, Ichinose M, Miyano M, Mimura K, Nishide I, Zhang BX, Wang SW, Zhou SO, Li BH. Mutations in hepatitis B virus core regions correlate with hepatocellular injury in Chinese patients with chronic hepatitis B. World J Gastroenterol 2005; 11(30): 4693-4696
- URL: https://www.wjgnet.com/1007-9327/full/v11/i30/4693.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i30.4693
Individuals infected with hepatitis B virus (HBV) may display asymptomatic, acute, fulminant, or chronic hepatitis. Previous studies have suggested that liver disorders due to HBV are immune-mediated and that hepatitis B envelope antigen (HBeAg) and hepatitis B core antigen (HBcAg) could be immunological targets[1-3]. Furthermore, several antigenic regions have been identified in the HBV core region using recombinant antigens or synthetic peptides. Some reports have revealed that amino acids (aa) 78-83 and aa 127-133 are exposed on the surface of HBcAg[4,5]. Others have also reported that aa 120-140 are related to the recognition of helper T-cells, and forms of the HBV core and e antigens influence cytokine release in mouse models[6-8]. Thus, liver disorder in hepatitis B is influenced by the host immune system, which attacks core peptides as the main target. Given this, the pattern of amino acid changes in the HBV core region might predict the clinical course according to the level of serum ALT (sALT) values and the presence of HBeAg. In other words, mutations in the HBV core region may help to predict the outcome of liver disorders. To identify one cause of hepatitis B activity, we analyzed the HBV core sequence and the frequency of core mutations.
Serum samples were taken from 17 Chinese patients, who were consistently positive for hepatitis B surface antigen (HBsAg). HBsAg and anti-hepatitis B surface antibody (HBaAb) were determined by immunoassay, while HbeAg, anti-hepatitis B envelope antibody (HBeAb), and HBcAb were determined by passive hemagglutination (PHA) assay. Patients were first divided according to the presence of HBeAg into positive (eAg-Po) and negative (eAg-Ne) groups, and then divided, according to the sALT values measured twice or thrice a year into high activity (HA) and low activity (LA) groups. The HA group had sALT over 100 IU/L at least once a year and the LA group had sALT consistently below 100 IU/L. This study was approved by the ethics committee and informed consent was obtained from all patients.
HBV DNA was extracted from 100 μL of serum. In brief, after the serum samples were diluted to 400 μL with distilled water, the DNA was extracted with 1 mL of phenol/chloroform (1/1:v/v) and precipitated with 1/10 volume of 3 mol/L sodium acetate and 2 volumes of 100% ethanol. The precipitates were dried under vacuum and dissolved with 20 μL of Tris-HCL and EDTA buffer (pH 7.4). PCR was performed to amplify a 609-bp DNA fragment using specific primers ( the 1st PCR primers : sense primer H: 5’-GGGAGGAGATTAGGTTA-3’, anti-sense primer I: 5’-GTACAGTAGAAGAATAAAG C-3’; /the 2nd PCR primers : sense primer D: 5’-CAAGCCTCCAAGCTGTG-CCT-3’, anti-sense primer F: 5’-ACCTTATG AGTCCA-AGGGAT-3’) corresponding to the outside of the core region in the HBV genome. A reaction mixture containing 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.0), 2.5 mmol/L MgCl2, 1 μmol/L each of the two primers, 200 μmol/L dNTP, 200 μg/mL gelatin, and 5 U of Taq DNA polymerase (Amersham Life Science, Cleveland, OH, USA) was added into a 200-μL tube containing 10 μL DNA solution. Amplification was performed for 35 cycles as follows: denaturation at 94°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 2 min. In the last cycle, incubation at 72°C was continued for 10 min to complete the extension. Electrophoresis of 10-μL aliquots of PCR products was performed on a 3.5 % agarose gel subsequently stained with ethidium bromide. To avoid contamination, we performed amplification under the stringent conditions recommended by Kwok and Higuchi, with one positive and one negative control for each sample[9].
After the primers were removed from the amplification products by the commercial product SUPREC-02™ (TAKARA, Otsu, Japan), the products were ligated to pT7BlueT vectors (Novagen Inc., WI, USA). The ligated phagemid vector was transfected into competent JM109 cells (Toyobo, Osaka, Japan). Several independent colonies were examined to find appropriate clones from PCR using D and F primers. At least three clones for each case were subjected to 2 mL of liquid culture and small-scale preparation of phagemid DNA. Each clone was reacted with the DyeDeoxy™ terminator cycle sequencing kit and sequenced with a DNA sequencing system (Model 373A, Perkin Elmer, Urayasu, Japan), and the consensus sequences were adopted. The amino acid (aa) sequences deduced from the DNA consensus sequences in the HBV core region were compared to the wild type, subtype adr[10] for all subjects.
Data values were presented as mean±SD. Statistical studies were achieved by Fisher’s exact probability test and unpaired Student’s t-test or Welch’s t-test. P < 0.05 was considered statistically significant.
Of the 17 patients in the study, 7 were in the eAg-Po group and 10 were in the eAg-Ne group. Similarly, 10 were in the HA group and 7 were in the LA group. The percentage of HBeAg/anti-HBe seroconversion was 80% in the HA group and 28.6% in the LA group. The percentage of females in HA group (50%) was greater than that in LA group (28.6%). More eAg-Po members were found in the HA group than in the LA group.
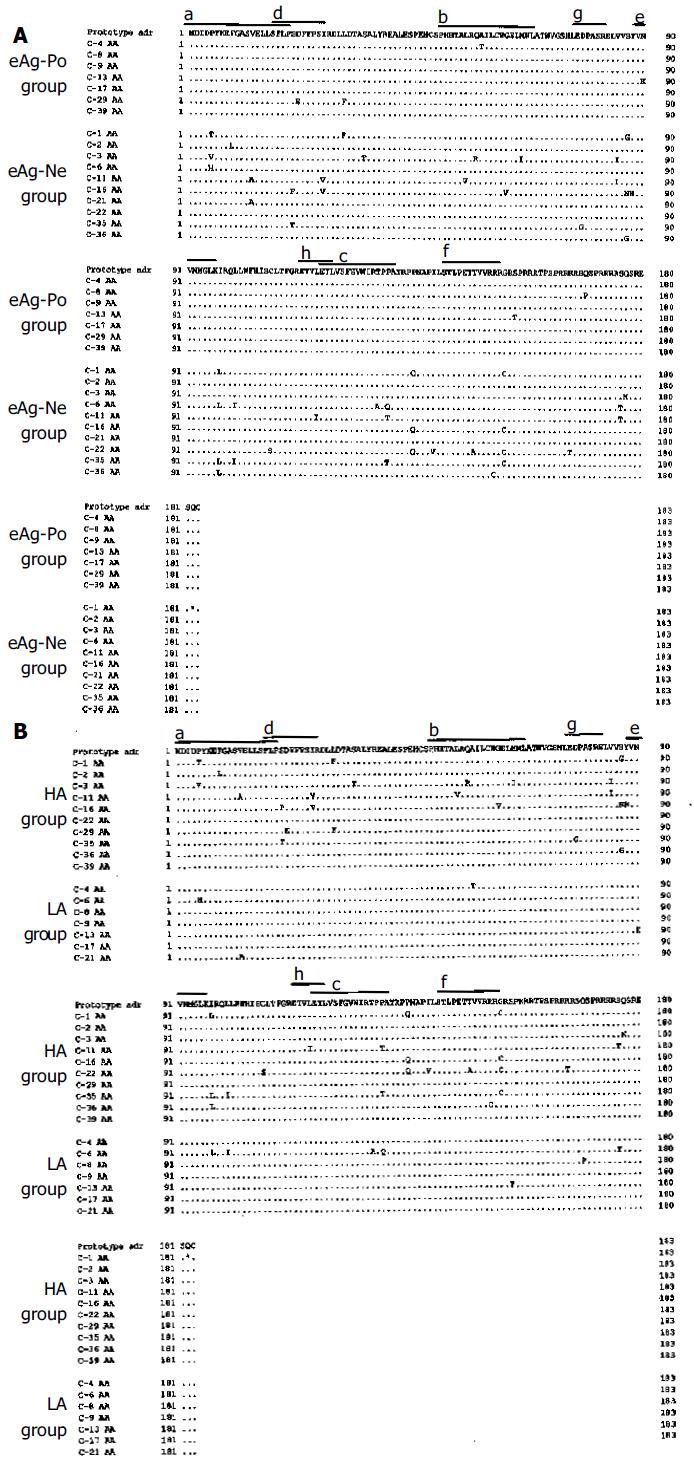
The DNA sequences in the HBV core region samples from 17 patients were analyzed and compared to the adr subtype (Figures 1A and B). Figure 1A shows the HBV core regions in the eAg-Po and eAg-Ne groups, whereas Figure 1B shows those in the HA and LA groups. As shown in Figures 1A and B, many amino acid changes were detected in the HBV core regions of the eAg-Ne group and the HA group. The frequency of DNA mutations and the amino acid changes in the HBV core region are summarized in Table 1. No significant difference was found in the number of DNA mutations between the LA and HA groups, whereas the number of DNA mutations in the eAg-Ne group tended to be higher than that in the eAg-Po group (P = 0.06). The number of amino acid changes in the HA group was significantly greater than that in the LA group (P < 0.05). The number of amino acid changes in the eAg-Ne group was also significantly greater than that in the eAg-Po group (P < 0.05).
As shown in Figures 1A and B, unique amino acid changes were detected in 30 of the 183 residues of the HBV core region. Of these 30 residues, six had an especially high mutation rate (more than 30%). The amino acid changes included replacements. Proline (Pro)-5 was replaced by threonine (Thr)-5, valine (Val)-5, and histidine (His)-5. Serine (Ser)-87 was replaced by glycine (Gly)-87 and asparagine (Asn)-87. Isoleucine (Ile)-97 was replaced by leucine (Leu)-97. Pro-130 was replaced by glutamine (Gln)-130 and Thr-130. Pro-135 was replaced by Gln-135 and Gly-153 was replaced by cysteine (Cys)-153.
Several virus factors and host factors have been reported to affect the activity of chronic hepatitis B[11-14]. In this study, we analyzed Chinese patients with chronic hepatitis B according to the level of sALT values and the presence of HbeAg, and then investigated the entire core region of HBV to characterize the amino acid changes in this region and to elucidate the relationship between the frequency of core mutations and the clinical activity of HBV-related liver diseases. We found that many amino acid changes in the HBV core region occurred in the HBV DNA of patients with high sALT values and with the seroconversion from HBeAg to anti-HBe. Increased activity of hepatitis and decreased time of seroconversion from HBeAg to anti-HBe may be attributed to increased immunological attacks against the HBV core region. Indeed, it has been reported that both B and T lymphocytes [helper T lymphocytes and cytotoxic T lymphocytes (CTL)] recognize core peptides. Some studies have reported that aa 78-83 and aa 127-133 are exposed on the surface of HBcAg[4,5]. Penna et al[15] and Ferrari et al[16] have identified three major HLA class II-restricted T cell recognition sites within HBcAg: aa 1-20, aa 50-69, and aa 117-131. Furthermore, it has been reported that the HLA-A2.1-restricted CTL epitope is mapped to aa 18-27, HLA-A31- and HLA-AW68-restricted-CTLs recognize aa 141-151[17], and HLA-A11-restricted-CTLs including HLA-A11 binding motifs recognize aa 88-96[18]. To our surprise, these recognition epitopes correspond to the HBV core region involving the most sites of amino acid changes, suggesting that the amino acid changes are not randomly distributed, but selectively generated. From these data, we can also suggest that core amino changes are influenced by the host immune system. Regarding the mechanism of the relationship between virus mutations and host immunity, it has been recently reported that amino acid changes in epitopes of T and B lymphocytes in the HBV core region may markedly influence T lymphocyte function or subsequent cytokine release. Therefore, we speculate that such amino acid changes affect the viral antigenicity against the host immunity and may be associated with the clinical severity of hepatitis. This study suggests that the frequency of core mutations may be associated with the severity of hepatitis and the HBV seroconversion from HBeAg to anti-HBe. On the other hand, we could not find any difference in the HBV core protein among several HBV viruses. To differentiate HBV among groups, it may be necessary to examine not only various HBV proteins including surface protein and polymerase protein, but also the type of HLA in the host. Further investigation is needed regarding other causes of activity in chronic hepatitis B.
We conclude that identifying mutations in the HBV core region is important for predicting the clinical course of hepatitis B.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 852] [Cited by in RCA: 861] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 2. | Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 377] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 3. | Ehata T, Omata M, Chuang WL, Yokosuka O, Ito Y, Hosoda K, Ohto M. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J Clin Invest. 1993;91:1206-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Pushko P, Sallberg M, Borisova G, Ruden U, Bichko V, Wahren B, Pumpens P, Magnius L. Identification of hepatitis B virus core protein regions exposed or internalized at the surface of HBcAg particles by scanning with monoclonal antibodies. Virology. 1994;202:912-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Sällberg M, Pushko P, Berzinsh I, Bichko V, Sillekens P, Noah M, Pumpens P, Grens E, Wahren B, Magnius LO. Immunochemical structure of the carboxy-terminal part of hepatitis B e antigen: identification of internal and surface-exposed sequences. J Gen Virol. 1993;74:1335-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Milich DR, Peterson DL, Schödel F, Jones JE, Hughes JL. Preferential recognition of hepatitis B nucleocapsid antigens by Th1 or Th2 cells is epitope and major histocompatibility complex dependent. J Virol. 1995;69:2776-2785. [PubMed] |
| 7. | Milich DR, Schödel F, Hughes JL, Jones JE, Peterson DL. The hepatitis B virus core and e antigens elicit different Th cell subsets: antigen structure can affect Th cell phenotype. J Virol. 1997;71:2192-2201. [PubMed] |
| 8. | Sällberg M, Rudén U, Wahren B, Noah M, Magnius LO. Human and murine B-cells recognize the HBeAg/beta (or HBe2) epitope as a linear determinant. Mol Immunol. 1991;28:719-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2583] [Cited by in RCA: 2475] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 10. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 769] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 11. | Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102-8110. [PubMed] |
| 12. | Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, Tsuda F, Tanaka T, Okamoto H, Miyakawa Y. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 213] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 13. | Inoue K, Yoshiba M, Sekiyama K, Okamoto H, Mayumi M. Clinical and molecular virological differences between fulminant hepatic failures following acute and chronic infection with hepatitis B virus. J Med Virol. 1998;55:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 14. | Peters M, Vierling J, Gershwin ME, Milich D, Chisari FV, Hoofnagle JH. Immunology and the liver. Hepatology. 1991;13:977-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 80] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Penna A, Bertoletti A, Cavalli A, Valli A, Missale G, Pilli M, Marchelli S, Giuberti T, Fowler P, Chisari FV. Fine specificity of the human T cell response to hepatitis B virus core antigen. Arch Virol Suppl. 1992;4:23-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Ferrari C, Bertoletti A, Penna A, Cavalli A, Valli A, Missale G, Pilli M, Fowler P, Giuberti T, Chisari FV. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J Clin Invest. 1991;88:214-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 171] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Missale G, Redeker A, Person J, Fowler P, Guilhot S, Schlicht HJ, Ferrari C, Chisari FV. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med. 1993;177:751-762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 173] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 18. | Tsai SL, Chen MH, Yeh CT, Chu CM, Lin AN, Chiou FH, Chang TH, Liaw YF. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD8+ cytotoxic T lymphocytes. J Clin Invest. 1996;97:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 2.1] [Reference Citation Analysis (0)] |