Published online Jan 21, 2005. doi: 10.3748/wjg.v11.i3.454
Revised: March 7, 2004
Accepted: April 13, 2004
Published online: January 21, 2005
AIM: To produce a recombinant protein rMBP-NAP, which was fusionally expressed by Helicobacter pylori (H pylori) neutrophil-activating protein (NAP) and E. coli maltose-binding protein (MBP) and to evaluate its immunoreactivity and immunogenicity.
METHODS: Neutrophil-activating protein gene of H pylori (HP-napA) was subcloned from the recombinant plasmid pNEB-napA, and fused to MalE gene of expressing vector pMAL-c2x. The recombinant plasmid pMAL-c2x-napA was confirmed by restriction enzyme digestion, and then transformed into E. coli TB1. Fusion protein rMBP-NAP was induced by IPTG and identified by SDS-PAGE analysis. Soluble rMBP-NAP was purified by amylose affinity chromatography. Immunoreactivity and immunogenicity of the fusion protein were evaluated by animal experiment, Western blotting with human H pylori anti-sera.
RESULTS: E.coli TB1 carrying recombinant plasmid pMAL-c2x-napA was constructed and led to a high efficiency cytosol expression of fusion protein rBMP -NAP when induced by IPTG. The molecular weight of rBMP-NAP was about 57 kD, accounting for 37.55% of the total protein in the sonicated supernatant of E. coli TB1 (pMAL-c2x-napA). The purity of the fusion protein after one-step affinity chromatography was 94% and the yield was 100 mg per liter of bacterial culture. The purified fusion protein could be specifically recognized by both human anti-sera from clinical patients with H pylori infection and rabbit sera immunized by rMBP-NAP itself.
CONCLUSION: Recombinant protein rMBP-NAP might be a novel antigen for vaccine development against H pylori.
- Citation: Kang QZ, Duan GC, Fan QT, Xi YL. Fusion expression of Helicobacter pylori neutrophil-activating protein in E.coli. World J Gastroenterol 2005; 11(3): 454-456
- URL: https://www.wjgnet.com/1007-9327/full/v11/i3/454.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i3.454
H pylori is now recognized as one of the most widespread human pathogens. Stomach mucosa colonized by H pylori is commonly accompanied with inflammatory infiltrates, consisting mainly of neutrophils and monocytes[1,2]. There is a good correlation between the degree of mucosal damage and neutrophil infiltration[3,4]. Several studies have provided evidence for the presence of protein components in H pylori water extracts capable of attracting and activating neutrophil adhesion to endothelial cells[5,6]. It is termed H pylori neutrophil-activating protein (HP-NAP). HP-NAP is localized in bacterial cytosol and released upon autolysis. It can bind to the external surface of the outer membrane, similar to what was found for urease[7,8]. In such a location, HP-NAP can mediate H pylori binding to the cell surface, thus, HP-NAP is supposed to be a major virulence factor and vaccine candidate[9]. To study the pathogenesis of H pylori infection and screen potential antigens for H pylori vaccine development, neutrophil-activating protein gene of H pylori (HP-napA) was amplified by PCR from H pylori MEL-HP27 strain, which was isolated from a clinical patient in Henan Province of China (preserved in Molecular Epidemiology Laboratory, Zhengzhou University). The napA sequence data have been published on GenBank (Accession No: AY366361) by the authors. Software analysis indicated that HP-napA gene was a highly conserved prokaryotic gene that could encode a 15-kD polypeptide. The aim of this study was to produce recombinant protein of HP-NAP in E.coli TB1 and to screen vaccine candidates. So the recombinant plasmid pMAL-c2x-napA was constructed, human recombinant mannose-binding protein (rMBP)-NAP expressed in E. coli TB1 was used as an antigen to immunize rabbits. Experiment with animal, H pylori human anti-serum and software were used to evaluate the immunogenicity and immunoreactivity of rMBP-NAP.
E.coli TB1, pMAL-c2x, amylose resin, and goat anti-rabbit IgG-HRP, goat anti-human IgG-HRP were purchased from NEB Company. pNEB-napA was constructed by the authors, 1 kb DNA ladder was from Sangon. Sal I, EcoRI, T4 DNA ligase, IPTG, Xgal were from Takara. Freund’s adjuvant was from Sigma; NC Western blot membrane was from Beijing Yili Company. DAB was from Roche Company, rabbits were from Laboratory Animal Center of Henan Province, H pylori human anti-sera were preserved in Molecular Epidemiology Department of Zhengzhou University.
Common DNA manipulation was done according to the methods described by Sambrook et al[10]. In brief, 0.5 μg pNEB-napA and 0.5 μg pMAL-c2x were digested respectively in 20 μL 1×H buffer with 1 μL SalI and 1 μL EcoR. Then, 2 μL pMAL-c2x digest and 1 μL napA fragment were mixed after purification, and 14 μL deionized H2O (Milli-Q) was added. DNA mixture was heated at 45 °C for 5 min and then cooled on ice, and 2 μL 10×T4 DNA ligase buffer and 1 μL T4 DNA ligase were added and incubated at 16 °C overnight.
The ligation was mixed with 25 μL competent TB1 cells, incubated on ice for 5 min, heated to 42 °C for 2 min. One hundred μL LB was added and incubated at 37 °C for 20 min, spread on a LB plate containing 100 mg/L ampicillin, incubated at 37 °C overnight. Colonies were picked with a sterile toothpick and inoculated onto a master LB amp plate and a LB amp plate containing 80 mg/L Xgal and 0.1 mmol/L IPTG. The colonies were incubated at 37 °C for 14 h and the white clones on LB amp plate containing Xgal and IPTG were selected. The corresponding patches on the master plate were determined. Plasmid DNA was miniprepared, and the insert of napA was identified by restriction enzyme digestion.
The positive clones of TB1(pMAL-c2x-napA) on the master plate were cultured in 5 mL broth containing amp at 37 °C, 200 r/min overnight. One hundred mL rich glucose broth containing amp was inoculated with 1 mL of the cultured TB1 cells. Cells of 2×108/mL(A600≈0.5) were cultured at 37 °C, IPTG was added to a final concentration of 0.3 mmol/L, and induced for 3 h. The expression of induced fusion protein was identified on SDS-PAGE. The cells were harvested and frozen at -20 °C overnight, sonicated in ice-water bath, centrifuged at 9000 g for 30 min at 4 °C. Soluble rMBP-NAP in the supernatant was purified by amylose affinity chromatography. The purity was evaluated by gel image analysis system (SynGene, USA) and the quantity was tested by Bradford assay.
Rabbits (3-5 kg) were immunized with purified rMBP-NAP four times. The primary immunization by hypodermic injection consisted of 100 μg rMBP-NAP and 0.5 mL complete Freund’s adjuvant. Three enhancement immunizations were performed 4 wk after the first injection, each dose consisting of 50 μg rMBP-NAP and 0.25 mL incomplete Freund’s adjuvant at weekly intervals. The sera were collected 3 wk after the last immunization. The negative sera were collected before the vaccination. Purified fusion protein was transferred to nitrocellulose (NC) membranes by semi-dry transfer cell (TRANS-BLOT.SD, BIORAD) at 15 V for 20 min after SDS-PAGE. Fusion protein impregnated NC strips were incubated with anti-rMBP-NAP rabbit serum (1:500) and anti-H pylori human serum (1:50) as the first antibody respectively, incubated with goat anti-rabbit IgG-HRP and goat anti-human IgG-HRP after washing. The secondary antibody was detected by reaction with DAB.
HP-napA and MBP-NAP gene sequence were inputted into Omega2.0 program. The sequences were translated into proteins and their antigenic features were calculated.
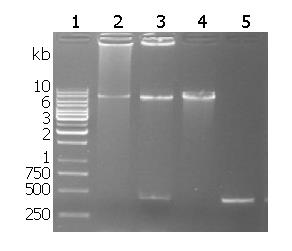
Recombinant plasmid restriction enzyme digestion is shown in Figure 1. The target fragment of 435 bp napA was inserted into the pMAL-c2x between EcoRI and sal I.
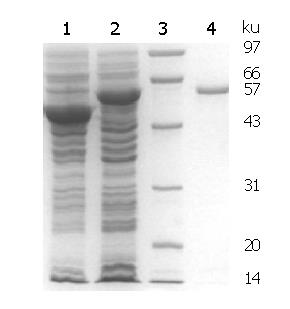
SDS-PAGE analysis of expressed and purified rMBP-NAP is shown in Figure 2. According to the gel image analysis system, the molecular weight of rMBP-NAP was about 57 kD, and the soluble fusion protein accounted for 37.55% of the total protein in the sonicated supernatant. The yield of fusion protein after affinity purification was 100 mg/L culture by Bradford assay.
Western blotting showed that the purified rMBP-NAP could be recognized with the sera from rabbits immunized with rMBP-NAP (Figure 3) and anti-H pylori sera from clinical patients infected with H pylori. In contrast, the result of the control sera from rabbits before vaccination and healthy people was negative.
H pylori infection is the major cause of chronic active gastritis and most peptic ulcer diseases[11]. It is also closely related to gastric cancers such as adenocarcinoma, mucosa-associated lymphoid tissue (MALT) lymphoma. Approximately half the population in the world is infected with H pylori. Vaccination might be the most effective and economic way to control this organism. Vaccine development is a process to identify the unique structures capable of generating immunological protection when formulated as a vaccine. So antigen and adjuvant selection is critical in developing H pylori vaccines. But so far the achievement in this area is still unsatisfied. Several studies have focused on identifying major virulence factors of H pylori such as CagA[12], urease and heat shock proteins. The toxicity of these natural proteins has limited their application and heat shock proteins might cause host immuno-cross reactions. A vaccine candidate must meet the following criteria: antigenic conservation among clinical isolates, elicitation of functional antibodies, protection of animal models, and ultimate safety and efficacy for human use [13]. So one of the tactical designs of subunit vaccines is to modify the structure of the virulence factors. Dundon et al reported that the majority of H pylori-infected patients were found to produce antibodies specific for HP-NAP, making it a strong vaccine candidate. We amplified HP-napA by PCR from H pylori MEL-HP27 strain and sequence analysis, and found that HP-napA gene was highly conserved among prokaryotic organisms. The results of this study showed that HP-napA fusionally expressed with maltose binding protein of E. coli did not degrade the immunoreactivity, but the immunogenicity of the fusion protein increased when the molecule was larger than natural HP-NAP. The feature of HP-NAP could be modified added with MBP. Moreover, maltose-binding protein provided a one-step affinity purification of the fusion protein using maltose resin, which could be cleaved from rMBP-NAP by factor Xa after purification. In conclusion, DNA recombinant technique and bioinformatics can provide an alternative approach to produce novel antigens for vaccine development. The features of rMBP-NAP as a vaccine candidate and the role of HP-NAP in H pylori-related pathogenesis should to be further evaluated in animal models.
Edited by Wang XL and Chen WW
| 1. | Dundon WG, Nishioka H, Polenghi A, Papinutto E, Zanotti G, Montemurro P, Del GG, Rappuoli R, Montecucco C. The neutrophil-activating protein of Helicobacter pylori. Int J Med Microbiol. 2002;291:545-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 2. | Nishioka H, Baesso I, Semenzato G, Trentin L, Rappuoli R, Del Giudice G, Montecucco C. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) activates the MAPK pathway in human neutrophils. Eur J Immunol. 2003;33:840-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Goodwin CS. Helicobacter pylori gastritis, peptic ulcer, and gastric cancer: clinical and molecular aspects. Clin Infect Dis. 1997;25:1017-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Bayerdörffer E, Lehn N, Hatz R, Mannes GA, Oertel H, Sauerbruch T, Stolte M. Difference in expression of Helicobacter pylori gastritis in antrum and body. Gastroenterology. 1992;102:1575-1582. [PubMed] |
| 5. | Montecucco C, de Bernard M. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 2003;5:715-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Namavar F, Sparrius M, Veerman EC, Appelmelk BJ, Vandenbroucke-Grauls CM. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect Immun. 1998;66:444-447. [PubMed] |
| 7. | Shimoyama T, Fukuda S, Liu Q, Nakaji S, Fukuda Y, Sugawara K. Helicobacter pylori water soluble surface proteins prime human neutrophils for enhanced production of reactive oxygen species and stimulate chemokine production. J Clin Pathol. 2003;56:348-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, Dunn BE. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905-912. [PubMed] |
| 9. | Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, Kelleher D, Rappuoli R, Montecucco C, Rossi F. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med. 2000;191:1467-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 236] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Sambrook J, Fritsch EF, Maniatis T. MOLECULAR CLONING: A Laboratory Manual. 2nded. Cold Spring Harbor Laboratory Press 1989: 888-897. . |
| 11. | Bai Y, Li LR, Wang JD, Chen Y, Jin JF, Zhang ZS, Zhou DY, Zhang YL. Expression of Helicobacter pylori Hsp60 protein and its immunogenicity. World J Gastroenterol. 2003;9:2711-2714. [PubMed] |
| 12. | Evans DJ, Evans DG. Helicobacter pylori CagA: analysis of sequence diversity in relation to phosphorylation motifs and implications for the role of CagA as a virulence factor. Helicobacter. 2001;6:187-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Chakravarti DN, Fiske MJ, Fletcher LD, Zagursky RJ. Application of genomics and proteomics for identification of bacterial gene products as potential vaccine candidates. Vaccine. 2000;19:601-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |