Published online Aug 7, 2005. doi: 10.3748/wjg.v11.i29.4511
Revised: January 1, 2005
Accepted: January 5, 2005
Published online: August 7, 2005
AIM: Eph receptors and ephrin ligands play a pivotal role in development and tissue maintenance. Since previous data have indicated an involvement of ephrin-B2 in epithelial healing, we investigated the gene expression and downstream signaling pathways induced by ephrin-B mediated cell-cell signaling in intestinal epithelial cells.
METHODS: Upon stimulation of ephrin-B pathways in IEC-6 cells with recombinant rat EphB1-Fc, gene expression was analyzed by Affymetrix’ rat genome 230 high density arrays at different time points. Differentially expressed genes were confirmed by real-time RT-PCR. In addition, MAP kinase pathways and focal adhesion kinase (FAK) activation downstream of ephrin-B were investigated by immunoblotting and fluorescence microscopy.
RESULTS: Stimulation of the ephrin-B reverse signaling pathway in IEC-6 cells induces predominant expression of genes known to be involved into wound healing/cell migration, antiapoptotic pathways, host defense and inflammation. Cox-2, c-Fos, Egr-1, Egr-2, and MCP-1 were found among the most significantly regulated genes. Furthermore, we show that the expression of repair-related genes is also accompanied by activation of the ERK1/2 MAP kinase pathway and FAK, two key regulators of epithelial restitution.
CONCLUSION: Stimulation of the ephrin-B reverse signaling pathway induces a phenotype characterized by upregulation of repair-related genes, which may partially be mediated by ERK1/2 pathways.
- Citation: Hafner C, Meyer S, Hagen I, Becker B, Roesch A, Landthaler M, Vogt T. Ephrin-B reverse signaling induces expression of wound healing associated genes in IEC-6 intestinal epithelial cells. World J Gastroenterol 2005; 11(29): 4511-4518
- URL: https://www.wjgnet.com/1007-9327/full/v11/i29/4511.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i29.4511
Eph receptor tyrosine kinases (RTKs) and their receptor-like ligands, the ephrins, represent the largest family of RTKs and are specialized on directing coordinated cell migration in development and possibly in tissue repair[1,2]. Based on their sequence homology, structure, and binding affinity, the Eph RTKs and ephrins are divided into two subclasses A and B. Usually, EphA receptors interact with ephrin-A ligands and EphB receptors with ephrin-B ligands. Both the receptors and the ligands are membrane-bound. A-ephrins are tethered to the outer leaflet of the plasma membrane by virtue of a glycosyl-phosphatidylinositol anchor, whereas B-ephrins are transmembrane proteins[3,4]. As a unique feature, upon receptor-ligand interaction, both receptor mediated “forward” and a “reverse” cellular signaling response are induced. Therefore, Eph/ephrin signaling represents a bidirectional cell-cell signaling system with ephrin ligands taking over receptor-like functions[5].
The rapid restitution of the intestinal surface after disruption of the epithelial barrier relies on the highly adaptive ability of epithelial wound-edge cells to rapidly form pseudopodial protrusions, reorganize the cytoskeleton, and migrate into a wound defect in a coordinated manner[6-8]. This fast epithelial healing is particularly important considering the necessity to protect the host against a considerable microbial threat and exposure to a multitude of immunogenic and toxic factors present in the gut[6].
Recent data suggest a pivotal role of Eph/ephrin signaling in tissue repair and maintenance of the gut. EphA2/ephrinA1 signaling has already been suggested to be involved in the homeostasis of the intestinal barrier in adults[9]. Moreover, differential expression of ephrin ligands and Eph-RTKs along the crypt-villus axis determines the correct positioning and allocation of the proliferating vs differentiating compartment during development of the intestinal crypts[10]. Interestingly, knockout of EphB2/B3 in mice leads to intermingling of both compartments and also displacement of Paneth cells to the top of the villi. More recently, the expression of a variety of Eph receptors and ephrin ligands has been demonstrated in the adult human gut by our group, with most prominent expression of EphA2, EphB2, ephrin-A1 and ephrin-B1/2[11]. Considering the important role of Eph receptors and ephrins development and their life-long highly organized expression in gut epithelium, a role in epithelial maintenance and repair seems possible. As a further step to understand their role in intestinal physiology, we investigated gene expression profiles in response to the stimulation of the ephrin-B dependent signaling pathways. Since many links between ephrin-B2 and MAP kinase (ERK) as well as focal adhesion kinase (FAK) mediated pathways have already been demonstrated[12,13], these pathways were also analyzed in more detail.
Non-transformed rat intestinal IEC-6 cells[14] were cultured in Dulbecco’s MEM (Biochrom AG, Berlin, Germany) supplemented with 5% fetal calf serum (FCS PAN Biotech, Aldenbach, Germany). After reaching 80% confluence, cells (passage 6) were starved overnight with 0.1% FCS and stimulated with 0.5 μg/mL (-3.3 nmol/L) rat recombinant EphB1-Fc (R&D Systems, Minneapolis, USA). Doses were determined according to the efficient stimulation of intestinal epithelial wound healing in scratch wound assays. Cells were harvested at 0 (control), 30, and 120 min in RLT buffer (Qiagen, Hilden, Germany). After homogenization, RNA isolation for further array analysis was performed using the RNeasy Mini Kit (Qiagen). RNA quality was assessed with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, USA) and quantity was measured spectrophotometrically.
For investigation of downstream transcriptional responses of EphB1-Fc stimulated pathways in IEC-6 cells, gene expression was analyzed by the Affymetrix rat genome expression set 230A (version 2.0). Sample preparation for microarray hybridization was carried out as described in the Affymetrix®GeneChip Expression Analysis Technical Manual. Briefly, 15 μg of total RNA were used to generate double-stranded cDNA (Invitrogen). Synthesis of Biotin-labeled cRNA was performed using the BioArrayTM HighYieldTM RNA Transcript Labeling Kit (ENZO Diagnostics). The length of the cRNA and fragmentation was confirmed using the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, USA). Affymetrix Microarray Suite (MAS) 5.1 was used for Single Array and Comparison Analysis. A global scaling strategy was applied, setting the average signal intensity of all arrays to a target value of 100. For RNA quality control and stringent data evaluation, the quality standards were applied as defined at a regional German Affymetrix Core Facility and Service Provider, the “CFB” (Competence Centre for Fluorescent Bioanalytics, http://www.kfb-regensburg.de).
Analysis of 331 regulated genes with changed P-values lower than 0.003 and higher than 0.997 (fold change >1.5) by the Ingenuity Pathway Analysis Software tool was used to get a better defined concept of the complex downstream events.
To confirm the differential expression of a selection of more than twofold regulated genes, real-time TaqMan RT-PCR (PE Applied Biosystems, Darmstadt, Germany) was performed on an ABI Prism 7900 HT Sequence Detection System as published elsewhere[11]. Briefly, cDNA was synthesized using the Reverse Transcription Kit from Promega (Madison, USA) according to the manufacturer’s protocol. Rat primers and probes for Cox-2, Egr-1/2, c-Fos, and monocyte chemotactic protein 1 (MCP-1) were obtained as Assays on Demand (Applied Biosystems). The standard curve method was used for the quantification of the relative amounts of gene expression products. This method provides unit less normalized expression values that can be used for direct comparison of the relative amounts of target mRNA in different samples[11]. All reactions were performed as triplicates. Probes and primers for TaqMan analysis for ephrin-B1 and -B2 were designed on the basis of gene-specific non-homologous DNA sequence of the corresponding members[11]. The sequences were as follows: ephrin-B1 (MGB probe TGTACTGGCTTGGGCC, forward CCTCC CCAGGCTTTGTGA, reverse TCCTGGCTGACCACA TCGT), ephrin-B2 (MGB probe TGCTCAGCGCTTAAA, forward GATGTGAAATTCAT-TTGTGGCAAT, reverse CCAGAAGTAGCTGTCA-ATTTGTTT).
IEC-6 cells were plated on fibronectin coated Lab-Tek chamber glass slides (Nalge Nunc Int., Naperville, USA) (100 000 cells/slide) and grown in Dulbecco’s MEM plus 5% FCS overnight. After washing with PBS, cells were fixed with 4% paraformaldehyde and after repeated wash steps with PBS blocked with 3% H2O2/CH3OH. Cells were again washed with PBS and incubated with Super Block (Zytomed, Berlin, Germany). For immunocytochemical staining, the following antibodies were used (30 min, 37 °C): goat EphB2 (1:100), rabbit ephrin-B1 (1:100), and rabbit ephrin-B2 (1:75) (Santa Cruz, CA, USA). Cells were incubated with Anti-Broad Spectrum Biotinylated antibody for 15 min (37 °C), and ZytoChemPlus HRP solution was added for 10 min at room temperature (Zytomed). After washing with PBS, the slides were stained with AEC chromogen for 10 min at room temperature (Dako, Hamburg, Germany). The reaction was stopped with H2O and counterstained with hemalaun. For negative control, the protocol was performed as mentioned above, but primary antibodies were omitted.
IEC-6 cells were grown in Dulbecco’s MEM (Biochrom AG, Berlin, Germany) supplemented with 5% FCS to 80% confluence. Cells were starved overnight with 0.1% FCS, then stimulated with 0.25 μg/mL (-1.65 nmol/L) rat recombinant EphB1-Fc (R&D Systems, Minneapolis, USA) and harvested at 0, 5, 10, 20, 30, and 45 min. Protein was isolated from the cells and the protein lysates were analyzed by SDS-PAGE and blotted according to standard protocols.
Activated (phosphorylated) FAK and pan-FAK were detected by the anti-(phosphotyrosine)-pY397 FAK antibody from Sigma (St. Louis, USA) and anti-FAK from Biosource (Camarillo, CA, USA). For detection of MAP-kinase phosphorylation, the following antibodies were used: anti-ERK1/2 (pT185, pY187) 1:1 000, anti-JNK1/2 (pT183, pY185) 1:2 500, anti-p38 (pT180, pY182) 1:1 000 (Biosource, Camarillo, CA, USA). Rabbit polyclonal anti-protein disulfide isomerase (PDI) antibody (dilution 1:4 000) served as a control for equal protein load (StressGen, Victoria, Canada). As secondary antibody, the anti-Rabbit-HRP (Cell signaling, Beverly, MA, USA) was used.
IEC-6 cells were plated on fibronectin coated glass cover slips and grown for 16 h in Dulbecco’s MEM plus 5% FCS. The monolayer was starved with Dulbecco’s MEM plus 0.1% FCS overnight, wounded by a cell scraper and stimulated with 0.5 μg/mL EphB1-Fc (-3.3 nmol/L). IgG-Fc (R&D Systems, Minneapolis, USA) served as a control. Cells were fixed with 4% paraformaldehyde for 5 min at room temperature and permeabilized with 0.1% Triton-X 100 for 5 min at 37 °C. After washing with PBS, cells were blocked with 1% bovine serum albumin for 15 min at 37 °C. For detection of activated FAK, an anti-(phosphotyrosine)-pY397 antibody (Sigma, St. Louis, USA) was used as primary and an anti-rabbit FITC-labeled antibody as secondary (Vector, Burlingame, USA). Cell nuclei were stained with DAPI (Chemicon, Temecula, USA). Images from the wound margin were taken on a Leitz microscope equipped with a fluorescence lamp and adequate filters. Cells were photographed at 400 magnification.
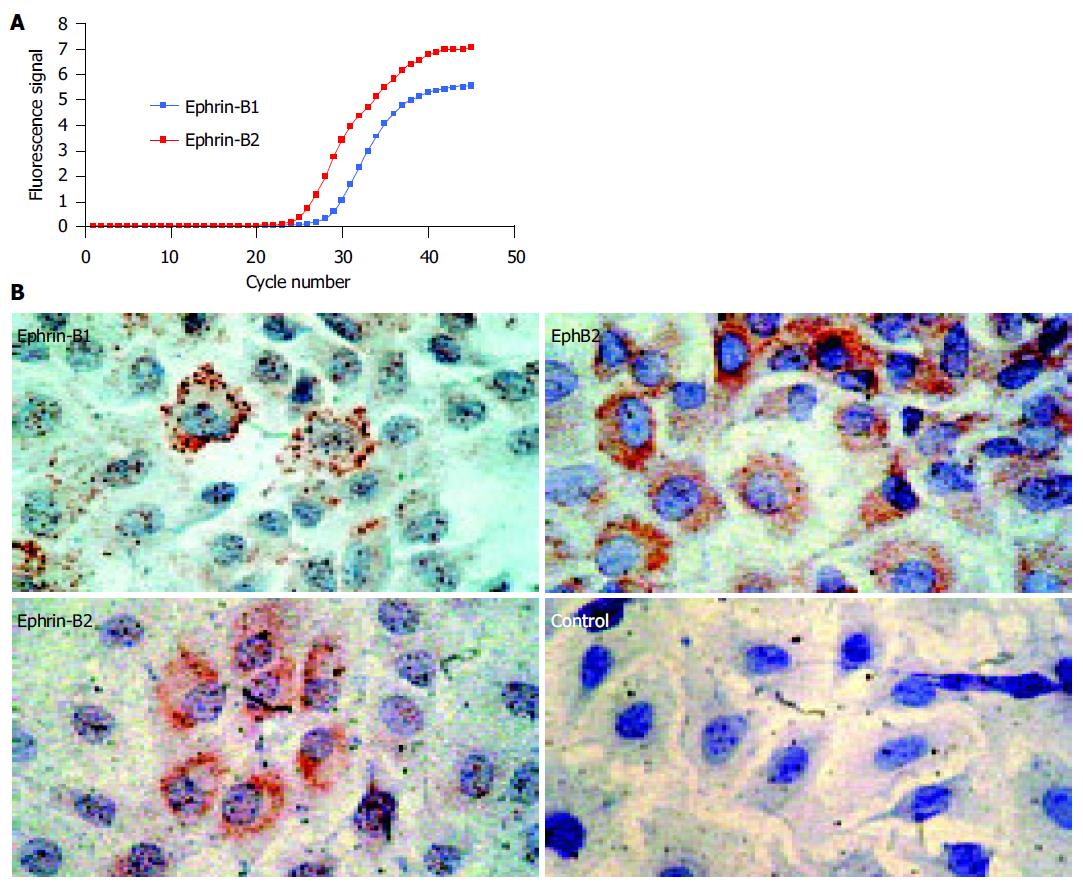
In the human small intestine and colon, coexpression of EphB2 and ephrin-B1/2 ligands has previously been demonstrated by our group[11]. To evaluate the suitability of IEC-6 cells as a model for investigation of EphB/ephrin-B dependent cell-cell signaling, the expression of both EphB2 receptor and ephrin-B1/2 ligands in IEC-6 cells was investigated. Real-time TaqMan RT-PCR revealed mRNA expression of ephrin-B1 and, more abundantly, ephrin-B2 in IEC-6 cells (Figure 1A). In addition, the expression of the EphB2 receptor and two corresponding high-affinity ligands in IEC-6 cells could be confirmed on the protein level by immunocytochemistry (Figure 1B). Hence, comparable to the human in vivo expression[11], IEC-6 cells coexpress the EphB2 receptor and two ephrin-B ligands in vitro. Therefore, cell-cell signaling between these cells seems possible.
Since previous investigations showed that ephrin-B mediated ’reverse’ signals generated upon binding of EphB receptors can specifically enhance intestinal wound healing in vitro, we were interested in the “downstream” target genes of the corresponding signaling cascades. Therefore, the transcriptional response to ephrin-B reverse signaling induced by recombinant EphB1-Fc was analyzed on the Affymetrix platform. The cells were stimulated with 0.5 μg/mL (-3.3 nmol/L) rat recombinant EphB1-Fc for 30 and 120 min. To identify the genes with highest possible probability to be truly regulated in these experiments, only genes were collected with significant detection-P values (exclusion of any false positives due to mismatch hybridization) and significant change-P values according to Affymetrix® algorithms. In addition, a minimum twofold change was set as cut-off. Table 1 shows the list of significantly regulated candidate genes after applying these stringent rules. Most of the genes are “early responders”, i.e., detectable after 30 min. However, a few of those genes were differentially regulated still after 120 min. Based on the current knowledge on gene function annotated to individual probe sets of the Affymetrix® rat chip, the regulated genes fell into seven major categories: Genes with direct links to (I) wound healing, (II) cell migration, (III) wound edge activation, (IV) pleiotrope transcription factors which also can directly participate in cell migration and wound healing, (V) genes that are functionally involved in apoptotic pathways, (VI) genes linked to host defense and inflammation, and (VII) genes that have host defense functions such as osmoregulation and detoxification (Table 1).
| Gene bank accession | Title | Function | FC (30 min1) | FC (120 min1) |
| gb:AA944459 | Dynein, cytoplasmic, light intermediate chain 1 | Migration/polarity/Wound edge | -2 | NC |
| gb:AA875047 | JQ0866 T-complex protein 1 – rat | Chaperonin/protein folding | -2 | NC |
| gb:AI231350 | Transcribed sequences | Unknown | -2 | NC |
| gb:AI071071 | Transcribed sequences | Unknown | -2 | NC |
| gb:BE097926 | Transcribed sequences | Unknown | -2.1 | NC |
| gb:AA925921 | ELV4_RAT ELAV-like protein 4 | Binds to the AU-rich element in c-fos | -2.1 | NC |
| and interleukin-3/mRNA degradation | ||||
| gb:AW144020 | Transcribed sequences | Unknown | -2.1 | NC |
| gb:U30789.1 | Upregulated by 1,25-dihydroxyvitamin D-3 | Unknown | -2.3 | -2.8 |
| gb:AI230596 | Importin alpha S1 | Nuclear transport/DNA binding | -2.3 | NC |
| gb:BF389856 | Beta-galactosidase, alpha peptide | Metabolism | -2.3 | NC |
| gb:NM_012903.1 | Acidic nuc. phosphoprotein 32 family, member A | Apoptosis | -3.2 | NC |
| gb:BF415939 | c-fos | Wound healing | 16 | NC |
| gb:NM_053633.1 | Early growth response 2 | Apoptosis/EGR2 induces apoptosis | 7.5 | NC |
| gb:NM_024360.1 | HES-1/hairy and enhancer of split 1 (Drosophila) | Notch pathway target/neurite | 4.9 | NC |
| outgrowth/morphogenesis | ||||
| gb:NM_012551.1 | Early growth response 1 | Apoptosis/EGR2 induces apoptosis | 4.6 | NC |
| gb:U03389.1 | Cox-2/prostaglandin-endoperoxide synthase 2 | Epithelial wound healing | 4.6 | 4 |
| gb:BE110108 | MAP kinase phosphatase-1/protein | Signaling/stress | 3.5 | NC |
| tyrosine phosphatase | ||||
| gb:NM_021836.1 | Jun B proto-oncogene | Transcription factor/wound healing cornea | 3 | NC |
| gb:AI178746 | Transcribed sequences | Unknown | 3 | NC |
| gb:NM_012620.1 | PAI-1/serine (or cysteine) proteinase | Affect wound healing by regulating the | 2.8 | NC |
| inhibitor, member 1 | fibrinolytic environment | |||
| gb:AI710284 | Beta-galactosidase, alpha peptide | Unknown | 2.8 | 3.2 |
| gb:BF420059 | Immediate-early protein pip92 | Unknown | 2.6 | NC |
| gb:BI288619 | c-jun homolog/v-jun sarcoma virus 17 | Transcription factor/wound repair/epithelial | 2.6 | NC |
| oncogene homolog (avian) | migration | |||
| gb:BI298889 | RBBP-2 | Transcription factor/cell cycle inhibition | 2.6 | NC |
| gb:NM_031642.1 | Core promoter element binding protein | Unknown | 2.5 | NC |
| gb:AB025017 | Zinc finger protein 36 | Unknown | 2.5 | NC |
| gb:AI599423 | DNA-damage-inducible protein GADD45 gamma | Cell cycle control | 2.5 | NC |
| gb:U02553.1 | MAP kinase phosphatase-1/protein | Signaling/stress | 2.3 | NC |
| tyrosine phosphatase | ||||
| gb:L81174.1 | Ankyrin-like repeat protein | Endothelial activation | 2.1 | NC |
| gb:NM_017206.1 | Solute carrier family 6, member 6 | Protects IECs from osmotic stress | 2.1 | NC |
| gb:BM383427 | Gp130/Interleukin 6 signal transducer | Host defense/tight junction loosening/ | 2.1 | 2.3 |
| proinflammatory lipid production | ||||
| gb:AI179464 | Rattus norvegicus RM1 mRNA, partial sequence | Unknown | 2.1 | 3.2 |
| gb:NM_031530.1 | MCP-1/small inducible cytokine A2 | Monocyte chemotaxis/candidate locus for exp. EM | 2 | NC |
| gb:NM_022858.1 | HNF-3/forkhead homolog-1 | Transcription factor/development | 2 | 2.6 |
| gb:AA850780 | Transcribed sequences | Unknown | 2 | NC |
| gb:NM_012940.1 | Cytochrome P450, subfamily 1B, polypeptide 1 | Detoxification | 2 | 5.3 |
| gb:NM_013057.1 | Coagulation factor 3 | Blood coagulation | 2 | NC |
| gb:BI284349 | Myeloid differentiation primary response gene 116 | Unknown | 2 | NC |
| gb:AI176519 | DIF-2/lipid responsive gene/Immediate | Differentiation | 2 | NC |
| early response 3 protein | ||||
| gb:AI169756 | G33_rat gene 33 polypeptide | Migration/signaling/adapter protein | 2 | NC |
| binds GTP-Cdc42/activates SAPK/JNK | ||||
| gb:BI288619 | c-jun homolog/v-jun sarcoma virus | Transcription factor/wound repair/epithelial | 2 | NC |
| 17 oncogene homolog (avian) | migration | |||
| gb:AA800192 | Rattus norvegicus endogenous | Unknown | 2 | NC |
| retrovirus mRNA, partial sequence | ||||
| gb:AW533292 | Similarity to protein ref: NP_073600.1 (H. sapiens) | Unknown | 2 | NC |
| gb:BF392456 | Rattus norvegicus non-erythrocyte | Unknown | 2 | NC |
| beta-spectrin mRNA, partial cds |
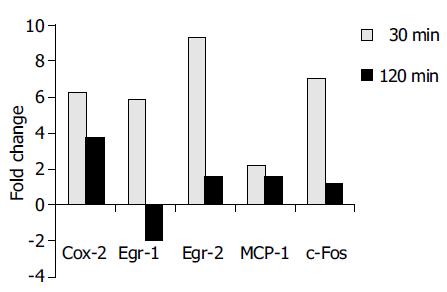
As real-time TaqMan RT-PCR has been proven to be a sensitive and reliable method for quantitative gene expression analysis[11], differential mRNA expression of selected candidate genes (c-Fos), cyclooxygenase 2 (Cox-2), early growth response gene 1 and 2 (Egr-1 and -2) and MCP-1 was confirmed using this method. The real-time RT-PCR results confirmed the array data very well (Figure 2). The c-Fos mRNA displayed a 7.1-fold upregulation after 30 min. Likewise, the strong induction of Egr-1, Egr-2, and Cox-2 after 30 min could be confirmed. In addition, RT-PCR revealed a 2.2-fold downregulation of Egr-1 mRNA after 120 min, which was not observed in the array data.
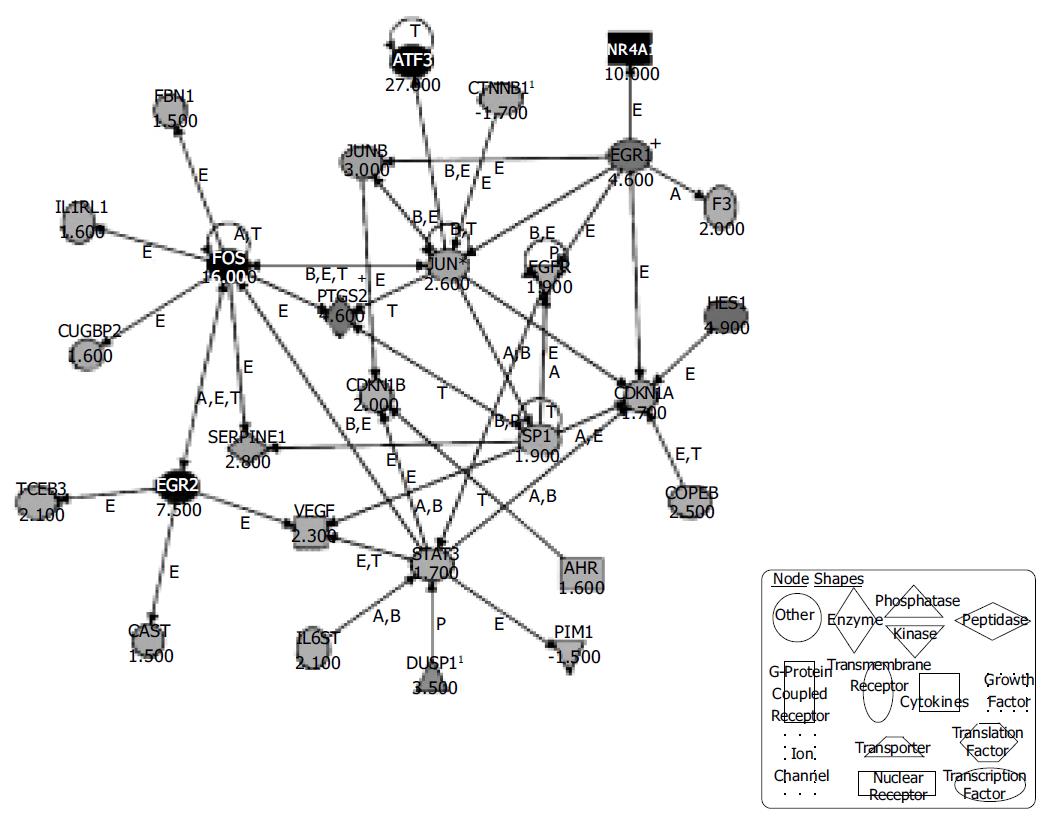
A total of 331 genes that were more than 1.5-fold regulated after 30 min of EphB1-Fc treatment and with MAS (Affymetrix Microarray Suite) change, P-values lower than 0.003 or higher than 0.997 respectively were used for the pathway analysis. The Affymetrix ProbeSet IDs were imported together with the fold changes into the Ingenuity Pathway Analysis Software (Ingenuity Systems). One hundred and thirteen genes were eligible for generating networks. All 21 networks found were ranked by score. The score is a numerical value, used to rank networks according to how relevant they are to the genes within the dataset. The network with the highest score contained 28 genes. A functional network of differentially regulated genes could be established including the confirmed candidates c-Fos, Egr-1 and Egr-2 together with further genes related to growth and apoptosis (Figure 3). Interestingly, the pathway tools revealed that the strongly upregulated genes Egr-2 and c-Fos are linked by mutual activation, expression and transcription of each other. Both Egr-1 and c-Fos interact with jun, which obviously has a central position in the demonstrated network. Furthermore, Egr-1 enhances the expression of the growth factor receptor EGFR and is further linked to the cell cycle via CDKN1A. The details of the conceptual ephrin-B dependent network are shown in Figure 3.
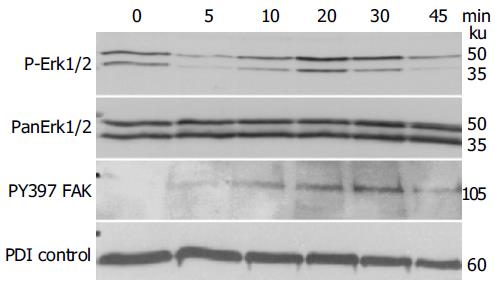
Previous work has shown that signaling responses to changes in the microenvironment regarding the presentation of Eph-RTKs or ephrins on neighboring cells involve major MAP kinase pathways such as JNK and Erk[13,15,16]. To identify the MAP kinase pathways involved in EphB/ephrin-B signaling in IEC cells, we screened for MAP kinase phosphorylation status in time-course experiments using stimulation with EphB1-Fc (0.25 μg/mL). As shown in Figure 4, there is a temporary induction of ERK1/2 phosphorylation that peaks after 20-30 min before returning to base levels.
Another important biological read-out of ephrin-B activation is the recruitment of FAK[2]. FAK is a crucial player in integrin-mediated attachment and migration as well as reorganization of focal adhesion complexes, which also protect from anoikis[17,18]. As shown in Figure 4, blotting with the anti-pY397-FAK indicates a time course of FAK phosphorylation upon EphB1-Fc stimulation in IEC-6 cells, which is almost identical to ERK 1/2 activation.
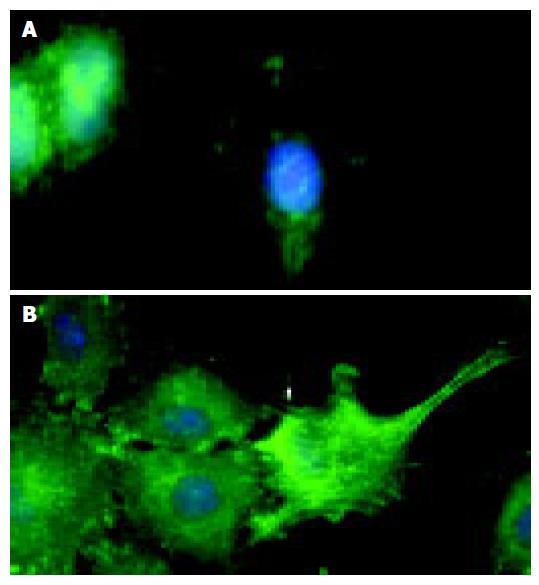
The latter observations could be further substantiated by immunofluorescence microscopy of wounded IEC-6 cell monolayers. Compared to IgG-Fc controls, EphB1-Fc stimulated IEC-6 cells revealed prominent induction of active pY397-FAK in neighboring cells at the wound margin (Figure 5).
Recent data generated by our group had suggested that EphB/ephrin-B cell-cell signaling can enhance epithelial wound closure of IEC-6 intestinal cells. In particular, the ephrin-B reverse signals seem to activate wound closure. In vivo, such functions could be essential for the protection of the host against microbial antigens and toxic factors abundantly present in the gut. In this context, it is also very interesting that we had observed significant imbalance of the ephrin-B ligand expression in intestinal epithelial cells isolated from patients with Crohn’s disease compared to healthy controls. Therefore, to elucidate further details of an assumed protective action of EphB-ephrin-B signal exchange in intestinal cells, we analyzed ‘downstream’ effects of the ephrin-B pathway. The most important finding of this study is that Affymetrix®® gene chip data revealed a significant upregulation of a set of genes known to be fundamentally involved into wound healing, e.g. c-Fos, Egr-1, Egr-2, and COX-2 (30 min after stimulation with recombinant EphB1-Fc). Egr-1 (4.6-fold) and c-Fos (16-fold) were among the most strongly early-induced genes. Egr-1 is known as a key activator of injury-induced gene expression[19]. Egr-1 might also be responsible for the observed induction of NR4A1 (immediate early gene transcription factor NGFI-B; 10-fold) and EGFR (epidermal growth factor receptor; 1.9-fold), as well as for the induction of jun B (3.0, fold) and c-jun (2.6, fold) according to the Ingenuity pathway analysis. In turn, the resulting AP-1 activity is known to be responsible for the observed prolonged induction of COX-2 (prostaglandin-endoperoxide synthase 2; 4.6-fold), but also MCP-1 (small inducible cytokine A2, 2.0-fold). COX-2 induction is considered as a further important line of defense for the gastrointestinal mucosa, necessary for maintenance of mucosal integrity and ulcer healing[20,21]. On the other hand, MCP-1 is a proinflammatory cytokine with a well defined role in inflammatory bowel disease[22-24]. Taken together, the Ingenuity pathway analysis suggests that stimulation of the ephrin-B signaling can induce a complex network of wound healing associated genes in intestinal epithelial cells with major interconnections to growth regulation and antiapoptosis, but also inflammation.
The exact anatomy of the signaling pathways activated in order to reseal epithelial defects in the gut remains poorly understood. IEC-6 cells, derived from fetal rat duodenal crypt cells, are one of the best-studied in vitro models for epithelial restitution[19]. Previous work on IEC-6 wounding has implicated the extracellular signal-regulated kinase (ERK), mitogen-activated protein kinase as a central mediator of wound-signal transduction and regulator of epithelial restitution. Monolayer injury results in rapid activation of ERK1/2 (p42 and p44) in IEC-6 cells[25]. Interestingly, our immunoblotting results are consistent with the view that ephrin-B signaling contributes to this pivotal wound-healing cascade. Moreover, in accordance with our data it has already been shown by another group that wounding of IEC-6 cells leads to ERK-dependent induction of Egr-1 and c-Fos. Both, direct blocking of ERK activation and interference with Egr-1 function resulted in diminished cell migration after injury[19]. This suggests that stimulation of ephrin-B is a part of the injury response program in intestinal epithelial cells involving ERK1/2, Egr-1, and c-Fos. Interestingly, activation of forward signaling by ligand-mediated stimulation of EphA2 and EphB2 receptors is also linked to the ERK/MAPK signaling cascade[26,27].
Recently, the receptor-like functions of B-ephrins and their intimate links to cell migration mechanisms could be defined in more detail on the molecular level[2,13]. Upon binding and dimerization of the cognate receptors of neighboring cells, the cytoplasmic c-terminus of ephrin-B undergoes phosphorylation due to the recruitment of Src-family kinases[15]. Subsequently, the phosphorylated cytoplasmic domain of ephrin-B provides a binding domain for SH2/SH3-adaptor proteins such as Grb4 (Nck2). In turn, Grb4 has three further SH3-domains that function as a scaffold for binding and activation of a variety of SH3-binding effectors. Some of them directly influence the dynamic control of the cytoskeleton, e.g., FAK, Paxillin, Abi-1, Axin, CAP, dynamin, hnRNPK, and Pak1[2,28]. Among those effectors, FAK is particularly interesting in this context, because active FAK is essential to control cell spreading, movements and also integrin-dependent survival of IEC[17,29]. Therefore, we analyzed FAK phosphorylation both by immunoblotting and by fluorescence microscopy. We show that in starved IEC-6 cells ephrin-B activation with nmol/L doses of EphB1-Fc leads to rapid induction of pY397-FAK, which seems to be most prominently detectable in wound-edge cells. The phosphorylation of FAK on Tyr397 is known to correlate with increased catalytic activity and appears to be important for the tyrosine phosphorylation of focal adhesion complex-associated proteins such as paxillin and Cas[17]. Theoretically, the directed turnover of such complexes finally determines the speed of cell movements and the capacity of an epithelium to reseal defects.
We conclude that ephrin-B signaling induces complex pathways, which are essential for rapid epithelial wound closure in the intestine. Functional links to major determinants of epithelial defense and coordinated cell movement place EphB/ephrin-B into the list of targets for future molecular therapies trying to intervene with such pathways, for instance in the context of inflammatory bowel disease. However, firstly the specifics of the side-effects such as the production of proinflammatory cytokines like MCP-1 and the complexity of seemingly counteracting downstream targets have to be investigated in more detail.
The skillful technical assistance and support of Mrs. Lydia Künzel and Mrs. Nadine Wandtke is gratefully acknowledged.
Co-first-authors: Christian Hafner and Stefanie Meyer
Co-correspondenct: Christian Hafner
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Drescher U. Eph family functions from an evolutionary perspective. Curr Opin Genet Dev. 2002;12:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Cowan CA, Henkemeyer M. Ephrins in reverse, park and drive. Trends Cell Biol. 2002;12:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 124] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Pasquale EB. The Eph family of receptors. Curr Opin Cell Biol. 1997;9:608-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 168] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Murai KK, Pasquale EB. 'Eph'ective signaling: forward, reverse and crosstalk. J Cell Sci. 2003;116:2823-2832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 284] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 5. | Holland SJ, Gale NW, Mbamalu G, Yancopoulos GD, Henkemeyer M, Pawson T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 419] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 6. | Wilson AJ, Gibson PR. Epithelial migration in the colon: filling in the gaps. Clin Sci (Lond). 1997;93:97-108. [PubMed] |
| 7. | Podolsky DK. Healing the epithelium: solving the problem from two sides. J Gastroenterol. 1997;32:122-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 128] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Jacinto A, Martinez-Arias A, Martin P. Mechanisms of epithelial fusion and repair. Nat Cell Biol. 2001;3:E117-E123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 283] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 9. | Rosenberg IM, Göke M, Kanai M, Reinecker HC, Podolsky DK. Epithelial cell kinase-B61: an autocrine loop modulating intestinal epithelial migration and barrier function. Am J Physiol. 1997;273:G824-G832. [PubMed] |
| 10. | Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 860] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 11. | Hafner C, Schmitz G, Meyer S, Bataille F, Hau P, Langmann T, Dietmaier W, Landthaler M, Vogt T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50:490-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 263] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 12. | Vindis C, Cerretti DP, Daniel TO, Huynh-Do U. EphB1 recruits c-Src and p52Shc to activate MAPK/ERK and promote chemotaxis. J Cell Biol. 2003;162:661-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Huynh-Do U, Vindis C, Liu H, Cerretti DP, McGrew JT, Enriquez M, Chen J, Daniel TO. Ephrin-B1 transduces signals to activate integrin-mediated migration, attachment and angiogenesis. J Cell Sci. 2002;115:3073-3081. [PubMed] |
| 14. | Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248-265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 651] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 15. | Palmer A, Zimmer M, Erdmann KS, Eulenburg V, Porthin A, Heumann R, Deutsch U, Klein R. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Mol Cell. 2002;9:725-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 228] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 16. | Huynh-Do U, Stein E, Lane AA, Liu H, Cerretti DP, Daniel TO. Surface densities of ephrin-B1 determine EphB1-coupled activation of cell attachment through alphavbeta3 and alpha5beta1 integrins. EMBO J. 1999;18:2165-2173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 162] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 17. | Parsons JT, Martin KH, Slack JK, Taylor JM, Weed SA. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene. 2000;19:5606-5613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 520] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 18. | Valentinis B, Reiss K, Baserga R. Insulin-like growth factor-I-mediated survival from anoikis: role of cell aggregation and focal adhesion kinase. J Cell Physiol. 1998;176:648-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Dieckgraefe BK, Weems DM. Epithelial injury induces egr-1 and fos expression by a pathway involving protein kinase C and ERK. Am J Physiol. 1999;276:G322-G330. [PubMed] |
| 20. | Halter F, Tarnawski AS, Schmassmann A, Peskar BM. Cyclooxygenase 2-implications on maintenance of gastric mucosal integrity and ulcer healing: controversial issues and perspectives. Gut. 2001;49:443-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 181] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 21. | Rodrigues S, Van Aken E, Van Bocxlaer S, Attoub S, Nguyen QD, Bruyneel E, Westley BR, May FE, Thim L, Mareel M. Trefoil peptides as proangiogenic factors in vivo and in vitro: implication of cyclooxygenase-2 and EGF receptor signaling. FASEB J. 2003;17:7-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 100] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Uguccioni M, Gionchetti P, Robbiani DF, Rizzello F, Peruzzo S, Campieri M, Baggiolini M. Increased expression of IP-10, IL-8, MCP-1, and MCP-3 in ulcerative colitis. Am J Pathol. 1999;155:331-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 216] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 23. | MacDermott RP. Chemokines in the inflammatory bowel diseases. J Clin Immunol. 1999;19:266-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 115] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | McCormack G, Moriarty D, O'Donoghue DP, McCormick PA, Sheahan K, Baird AW. Tissue cytokine and chemokine expression in inflammatory bowel disease. Inflamm Res. 2001;50:491-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 25. | Göke M, Kanai M, Lynch-Devaney K, Podolsky DK. Rapid mitogen-activated protein kinase activation by transforming growth factor alpha in wounded rat intestinal epithelial cells. Gastroenterology. 1998;114:697-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Pratt RL, Kinch MS. Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene. 2002;21:7690-7699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 109] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Tong J, Elowe S, Nash P, Pawson T. Manipulation of EphB2 regulatory motifs and SH2 binding sites switches MAPK signaling and biological activity. J Biol Chem. 2003;278:6111-6119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Cowan CA, Henkemeyer M. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature. 2001;413:174-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 259] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 29. | Ray RM, Viar MJ, McCormack SA, Johnson LR. Focal adhesion kinase signaling is decreased in polyamine-depleted IEC-6 cells. Am J Physiol Cell Physiol. 2001;281:C475-C485. [PubMed] |