Published online Jun 21, 2005. doi: 10.3748/wjg.v11.i23.3523
Revised: July 3, 2004
Accepted: August 30, 2004
Published online: June 21, 2005
AIM: To compare the distribution of virulence-associated genotypes of Helicobacter pylori (H pylori) in two areas of north China with different gastric cancer risk and furthermore probe into the pathogenicity of the bacterium.
METHODS: Gastric biopsies were taken from 355 subjects from Zhuanghe, a high risk area of gastric cancer, and 136 subjects from Shenyang, a low risk area of gastric cancer. A total of 149 H pylori strains isolated from these patients were studied by PCR for differences in the genotypes of cagA, vacA, and iceA.
RESULTS: In patients with high risk for gastric cancer, higher frequencies of vacA s1 or s1m1b genotypes were found as compared to those from the low risk area.
CONCLUSION: There is significantly different distribution of H pylori genotypes between Zhuanghe and Shenyang areas in north China.
-
Citation: Gong YH, Wang Y, Yuan Y. Distribution of
Helicobacter pylori in north China. World J Gastroenterol 2005; 11(23): 3523-3527 - URL: https://www.wjgnet.com/1007-9327/full/v11/i23/3523.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i23.3523
Helicobacter pylori (H pylori) colonizes the human stomach and establishes long-term infection of the gastric and duodenal mucosa[1]. Although H pylori infects approximately half of the world’s population, only a small proportion of infected subjects develop symptoms or clinically significant diseases. The reasons for this fact are unknown but may be related to the host’s immunological defenses, environmental factors and/or the virulence of different strains of the bacteria[2,3]. Several genes have been identified that may play a role in the pathogenicity of H pylori genotypes such as cagA, vacA, and iceA[4].
Many researches have shown H pylori genotype not only has a different disease association but also has a particular geographic distribution. CagA+ strains are more commonly associated with peptic ulceration, atrophic gastritis, and adenocarcinoma of the stomach than cagA- strains[5,6] in many northern countries. Subtype vacA s1a is predominant in populations of northern European ancestry, and is associated with duodenal ulcer. Subtype s1b is predominant in Africa and very frequently found in Portugal, Spain, and Central and South America. In France, Italy, and the USA, the frequency of s1a and s1b genotypes is similar[7]. In Guangdong and Zhejiang Provinces, and Shanghai of China, the main genotype is vacAs1m1[8,9]. IceA genotype also has a particular geographic distribution. IceA1 is predominant in Japan and Korea, iceA2 is predominant in America. In Columbia iceA2 is the predominant genotype in gastric cancer and gastritis. But the distribution of H pylori genotypes in north China is not reported.
Disease associations (e.g., with duodenal ulcers) have been proposed for the cag pathogenicity island (cagA for marker), vacA and iceA. However, these associations are not consistent in different geographic regions. Though genotyping of cagA, vacA, and iceA appears not to be useful for disease specificity in some regions, it may play a role in molecular epidemiological studies in terms of identifying the predominant H pylori strain that is circulating in a given geographic area.
Zhuanghe in Liaoning Province is a high-risk area of gastric cancer in north China. The mortality rate of gastric cancer is 50 per 100000 persons, compared to 14.5 per 100000 (1995) persons in Shenyang, a low risk area of north China[10]. The prevalence of H pylori infection in adults from Zhuanghe is more than 60%, compared to 12% from Shenyang. A previous research from the high-risk area in Zhuanghe reported that there is a significantly positive relationship between gastric diseases or precancerous lesions and H pylori infection[11], suggesting there may be some relationship between H pylori infection and gastric cancer incidence in these areas. But whether the high incidence of gastric cancer in the high-risk area has a relationship with specific H pylori genotype is still unclear. The aim of this study was to compare the distribution of H pylori genotypes in the two areas with different cancer risk and furthermore explore its geographic characteristics.
A total of 491 cases were involved in this study including 136 cases from Shenyang (69 men and 57 women, 25-78 years, mean age: 48.61 years), 355 cases from Zhuanghe (174 men and 181 women, 21-79 years, mean age: 49.33 years). Their biopsies were obtained during endoscopy with informed consent. One antrum biopsy was taken for culture and stored at -70 °C in 0.5 mL of brucella broth (Difco) with 15% glycerol until incubation. Three biopsies were taken for pathology diagnoses, from gastric antrum, corpus, and angularis, respectively.
Culture was prepared by smearing single biopsy specimens on petri plates containing brain heart infusion (BHI) agar (Difco) supplemented with 7% sheep blood, 0.4% IsoVitale X, amphotericin B (8 μg/mL), trimethoprim (5 μg/mL), and vancomycin (6 μg/mL) and incubated at 37 °C in an atmosphere of 5% O2-100 mL/L CO2-85% N2 for 3-6 d. H pylori colonies were identified based on their typical morphology, characteristic appearance on Gram staining, a positive urease test, and subsequent gene-specific PCR tests. H pylori cells that grew out from biopsy on the primary culture plate were collected as a pooled population, and preserved in sterile BHI broth with 15% glycerol at -70 °C. In general, only one such a culture was analyzed per patient.
The strain was centrifuged, the supernatant was removed, and then suspended in cell lysis, incubated at 37 °C overnight. The DNA was extracted with phenol-chloroformisoamyl alcohol by standard procedures and precipitated by the addition of 1/10 volume of ammonium acetate and 2.5 volume of cold ethanol. After centrifugation, the DNA pellet was washed with 70% ethanol and dissolved in TE buffer (10 Mm Tris-HCl [pH 8.3], 0.1 mmol/L EDTA).
The integrity of the DNA was assessed by 0.7% agarose gels stained with ethidium bromide. Polymerase chain reaction (PCR) was performed in a volume of 20 μL containing 10 pmoL of primer, 0.5 μL genomic DNA, 2.5 mmol/L of each of 4 dNTPs (Takara Company), and 2.5 U of Taq DNA polymerase (Takara Company). PCR amplifications were performed in an automated thermal cycler (Biometra Co., Germany). Table 1 summarizes the primer sequences and the expected size of PCR products. The following cyclical conditions were used: for vacA: 35 cycles of 1 min at 94 °C, 1 min at 52 °C, and 1 min at 72 °C; for cagA: 1 min at 94 °C, 1 min at 50 °C, and 1 min at 72 °C; for iceA: 1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C. The amplified PCR products were resolved in 2% agarose gels containing 0.5*TBE, stained with ethidium bromide and visualized under a short wavelength ultraviolet light source.
| Gene and DNA region | Prime | Primer sequence (5’-3’) | Size of PCR product (bp) | References |
| cagA | CAGAF | GGCAATGGTGGTCCTGGAGCTAGGC | 324 | Pan[21] |
| CAGAR | GGAAATCTTTAATCTCAGTTCGG | |||
| vacAs1 and s2 | VA1-F | ATGGAAATACAACAAACACAC | 259/286 | Atherton[22] |
| VA1-R | CTGCTTGAATGCGCCAAAC | |||
| m1a | VA3-F | GGTCAAAATGCGGTCATGG | 290 | Atherton[22] |
| VA3-R | CCATTGGTACCTGTAGAAAC | |||
| m1b | VAm-F3 | GGCCCCAATGCAGTCATGGAT | 291 | Atherton[23] |
| VAm-R3 | GCTGTTAGTGCCTAAAGAAGCAT | |||
| m2 | VA4-F | GGAGCCCCAGGAAACATTG | 352 | Atherton[22] |
| VA4-R | CATAACTAGCGCCTTGCAC | |||
| iceA1 | iceA1F | GTGTTTTTAACCAAAGTATC | 247 | Peek[10] |
| iceA1R | CTATAGCCASTYTCTTTGCA | |||
| iceA2 | iceA2F | GTTGGGTATATCACAATTTAT | 229/334 | Peek[10] |
| iceA2R | TTRCCCTATTTTCTAGTAGGT |
Data were analyzed using SPSS for windows version 11.0. The χ2 test or Fisher’s exact test was used to assess the relationships between different areas. P<0.05 was considered statistically significant.
A total of 149 H pylori strains out of 491 biopsies from two geographic areas in China were obtained. The high-risk group comprised 102 strains from Zhuanghe. The low risk group comprised 47 strains from Shenyang. cagA, vacA, and iceA genotypes of H pylori strains were analyzed by PCR. Genotyping results are summarized in Table 2.
| Gene | Zhuanghe area (%) n = 107 | Shenyang area (%) n = 42 |
| cagA+ | 101 (94.4) | 42 (100.00) |
| cagA- | 6 (5.6) | 0 (0) |
| vacA s1 | 102 (95.33)b | 27 (64.29) |
| vacA s2 | 0 (0) | 0 (0) |
| vacA s- | 5 (4.67)d | 15 (35.71) |
| vacA m1a | 0 (0) | 0 (0) |
| vacA m1b | 24 (22.43) | 5 (11.9) |
| vacA m2 | 26 (24.3) | 11 (26.19) |
| vacA m1b/m2 | 48 (44.96) | 22 (52.38) |
| vacA m- | 9 (8.41) | 4 (9.52) |
| iceA1 | 10 (9.35) | 1 (2.38) |
| iceA2 | 10 (9.35) | 1 (2.38) |
| iceA1/iceA | 2 84 (78.5) | 32 (76.19) |
| iceA- | 3 (2.8) | 8 (19.05) |
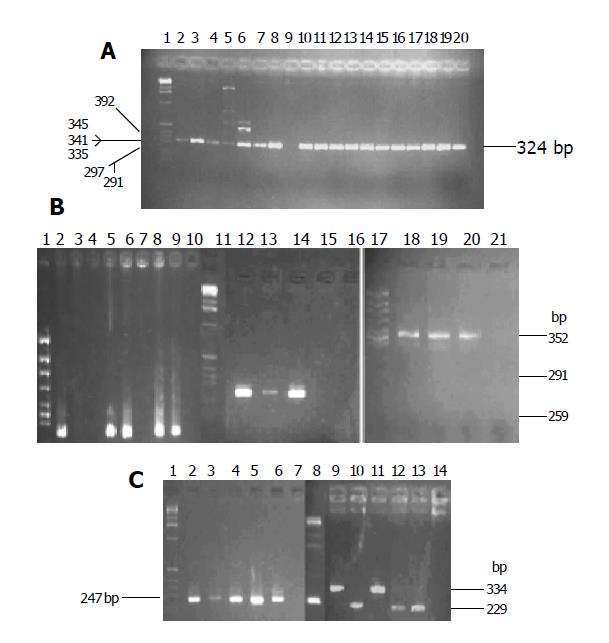
There was a high prevalence of the cagA+ strain in the two areas. The cagA gene was detected in 101 of 107 H pylori isolates in Zhuanghe (94.4%) and 42 of 42 in Shenyang (100%). There was no difference in the distribution between the two areas. Six strains did not yield any PCR product for cagA (Table 2 and Figure 1A).
Specific primer was used to type allele of the vacA gene. The prevalence of vacA s1 strains was significantly higher in patients from Zhuanghe (95.33%), as compared to those from Shenyang (64.29%, P<0.001). The positive isolate yielded a 259-bp PCR fragment characteristic of the s1 (potentially more virulent) allele, and none yielded the 286-bp fragment characteristic of the s2 (less virulent) allele in both areas (Figure 1B). Twenty strains did not yield any PCR product for vacA s (Table 2).
The prevalence of vacA m1b and m2 strains had no difference in both areas (22.43% vs 11.95%, 24.3% vs 26.195%, respectively, P>0.05). VacA m1a was not found in either area. Forty-eight and twenty-two strains were found, respectively in the high and low risk areas, m1b and m2 were detected in one patient at the same time. Fourteen strains of vacA m gene could not be genotyped (Table 2 and Figure 1B).
Based on analysis of the vacA s and m region, we examined vacA s1m1b, vacA s1m2, vacA s1m1b/m2, vacA s1m-, vacA s- different combinations in patients. The vacA genotype s1m1b was significantly higher in Zhuanghe than in Shenyang (20.56% vs 2.38%, P<0.01). The predominant vacA genotype in both areas was s1m1b/m2. However, the difference was not statistically significant (Table 3).
IceA gene could be genotyped as iceA1 and iceA2 with specific primers. The prevalence of iceA1 and iceA2 strains had no difference in both areas (9.35% vs 2.38%, 9.35% vs 2.38%, respectively, P>0.05). Eighty-four isolates (78.5%) in Zhuanghe and 32 isolates (76.19%) in Shenyang were positive for both iceA1 and iceA2, and 11 isolates (21.85%) did not yield any PCR product for iceA (Table 2 and Figure 1C).
We examined several different combinations based on the analysis of the vacA s region (s1 and s2) and m region (m1a, m1b, m2), cagA (positive and negative), and the iceA type (iceA1 and iceA2) in patients (Table 4). The predominant combination genotypes in both areas were cagA+, vacA s1/m1b/m2, iceA1/iceA2 (36.45% vs 33.3%, P>0.05, Table 5).
| cagA | vacAs | vacAm | iceA | Zhuanghe (n = 107) | Shenyang (n = 42) |
| cagA | s1 | m1b | A1 | 3 (2.8) | 0 (0) |
| cagA | s1 | m1b | A2 | 3 (2.8) | 0 (0) |
| cagA | s1 | m1b | A1+A2 | 13 (12.15) | 0 (0) |
| cagA | s1 | m2 | A1 | 3 (2.8) | 0 (0) |
| cagA | s1 | m2 | A2 | 1 (0.935) | 1 (2.38) |
| cagA | s1 | m2 | A1+A2 | 18 (16.82) | 5 (11.9) |
| cagA | s1 | m1b+m2 | A1+A2 | 39 (36.45) | 14 (33.3) |
| cagA | s | m1b+m2 | A1+A2 | 2 (1.87) | 7 (16.67) |
| Others | 25 (23.36) | 15 (14.02) | |||
| Genotype | Zhuanghe (n = 107) | Shenyang (n = 42) |
| vacA m1/m2 | 48 (44.86) | 22 (52.38) |
| iceA1/iceA2 | 84 (78.5) | 32 (76.19) |
| vacAm1/m2 or iceA1/iceA | 92 (85.98) | 33 (78.57) |
More than half of the people are infected with H pylori in the world, but not all individuals developed associated diseases[12]. This may be related to a complex of environmental factors, host characteristics and bacterial virulence determinants. Several virulence-associated factors of H pylori have been associated with clinical outcomes of the infection. In the early study we have reported that more than 60% individuals in the high-risk area of gastric cancer in Zhuanghe are infected with H pylori; and the infection has a significant association with gastric disease[11]. But the geographic characteristics of H pylori infection in north China have not been well described before. In the present research, we examined the distribution of H pylori genotypes in the high and low risk areas of gastric cancer in north China. The results indicate that vacA s1 or s1m1b genotype is more prevalent in Zhuanghe where gastric cancer incidence is high, whereas vacA s-genotype is relatively more frequent in Shenyang where gastric cancer incidence is very low. Vacuolating cytotoxin encoded by vacA gene can aggregate into flower-shaped hexamers and heptamers, which represent the mature active toxin. The mature toxin is peculiarly suited to the gastric environment because it is activated by acid and its activated form causes more profound epithelial changes[13]. Infection with H pylori possessing vacA s1 is associated with a higher degree of neutrophil and lymphocytic infiltration of the human gastric mucosa, and the presence of cagA[14]. In human stomach, strains with vacA m1 allele are associated with severe epithelial damage compared to those with m2 allele[15]. The different m types seem to recognize different receptors on epithelial cells and may induce different intracellular responses. Previous studies have shown that vacA s1m1 strains produce large amounts of vacuolating cytotoxin and that these strains are associated with peptic ulcer disease (PUD)[16]. On the other hand, the s2m2 strains produce no or only small amounts of cytotoxin and are uncommon in patients with PUD. The s1m2 strains seem to take an intermediate position[16]. Miehlke et al[17], reported that the vacA s1, m1 genotypes are more frequently detected in H pylori from gastric cancer (GC) patients (70.6%) than from mucous associated lymph tumor (MALT), duodenal ulcer (DU), and functional dyspepsia (FD) patients (P<0.05) and may be used to identify infected patients at an increased risk for GC. A recent study showed that, when cocultured with AGS gastric epithelial cells, H pylori strain 60190, which expresses s1m1 VacA toxin, induces significantly higher levels of apoptosis than isogenic vacA null mutant strain[18]. The risk for developing gastric cancer is >90-fold higher in patients with severe multifocal atrophic gastritis than in patients with normal mucous[19]. Thus, patients infected with vacA s1m1 strains have a higher risk of carcinogenesis. In our study, distribution of the vacA s1m1 genotype was found in the high and low risk areas, this may be a factor contributing to the higher gastric cancer incidence in the high-risk area. Some conclusions may be drawn from the fact that patients infected with this strain are the high-risk persons for gastric cancer.
In addition, we did not find vacA s genotype in 20 isolates. This is currently unexplained and may be due to the existence of additional vacA. Pan et al[20], reported that 78 of 96 H pylori isolated from Shanghai and Guangzhou carry m2, they thought m1b alleles seem infrequent in China. But in our study 24 (22.42%) isolates from Zhuanghe and 5 (11.95%) isolates from Shenyang carried the canonical m1b allele that has a similar prevalence to m2 allele (24.3% and 26.9%, respectively). The distinct distribution of vacA alleles in different regions of China suggests that H pylori distribution has significant geographical characteristics. The studies of isolates from patients in different countries or regions give only a partial view of H pylori as a globally distributed human pathogen.
Detailed molecular analyses[16,21] have shown that each H pylori strain only contains a single vacA s region, m region and iceA allele. VacA s1 and s2, m1 and m2, as well as iceA1 and iceA2 can be considered as mutually exclusive genotypes of a single strain. Consequently, if multiple genotypes are found, this is a strong indication of the presence of multiple strains. In our study, 48 cases in Zhuanghe (44.86%) and 22 cases in Shenyang (52.38%) showed evidence of multiple vacA genotypes. In the iceA locus, 84 cases (78.5%) and 32 cases (76.19%) in the high and low areas were positive for iceA1 and iceA2. Considering vacA and iceA genes, the presence of multiple H pylori strains was found respectively in 92 (85.98%) and 33 (78.57%) patients from the two areas. It may be speculated that more than one strain may be acquired in childhood, especially in countries with a very high prevalence of H pylori. It is not known whether multiple strains colonize simultaneously (co-infection) or at different time points (superinfection). The co-existence of more than one strain in the same individual may reflect the adaptation of multiple bacterial genotypes to different, non-overlapping microniches in the same stomach. The dynamics of co-colonization by multiple strains has been studied in animal models[22]. Non-human primates experimentally challenged with a mixture of strains show only a transient infection by more than one strain, and then a single strain becomes predominant over the others[22]. The early study from Mexico found there is some association between multiple strain infections and peptic ulceration[18]. One possible reason for such an association may be that multiple strain infection increases the chance of infection with a more pathogenic strain or mixed H pylori strains may act synergistically to persist in the stomach and cause damage[23]. Whether multiple strain infection has some practical significance in north China is to be discussed further.
Combined analysis of vacA, cagA, and iceA genotypes may permit identification of high-risk patients infected with more pathogenic H pylori strains. Eventually, patients infected with such strains could be selected for prophylactic anti-H pylori treatment to prevent associated gastric diseases later in life. For example, Figueiredo et al, reported that the s1/m1/cagA+/iceA1 and s1/m1/cagA+/iceA2 strains are more predominant in patients with duodenal ulcer (27.6% and 17.2%), gastric ulcer (50% and 37.5%) and gastric carcinoma (36.8% and 36.8%), whereas s2/m2/cagA-/iceA2 strains are predominant in patients with gastritis only (31.3%). Yamaoka et al[4], used PCR to examine iceA, vacA, and cagA status of 424 H pylori isolates obtained from patients of four different countries and found that the cagA+/iceA1/vacAs1cm1 genotype is predominant in Japan and Korea, the cagA+/iceA2/vacAs1bm1 genotype is predominant in the USA, and the cagA+/iceA2/vacAs1am1 genotype is predominant in Columbia. But there is no association between the iceA, vacA, or cagA status and clinical outcome. In our study, the predominant genotype in both high- and low-risk areas was cagA+/vacAs1m1bm2/iceA1/iceA2 and had no correlation with other reports. This may be ascribed to the high percent of multiple strain infection in both areas. Whether this result has some relationship with associated gastric diseases is to be discussed further.
In our study, the cagA gene was found in all the 42 isolates from Shenyang and 101 (94.4%) isolates from Zhuanghe. There was no difference in the distribution of H pylori genotypes in the two areas. This high prevalence contrasts with the 30% cagA- frequency of H pylori in Western countries. Bravo et al[12], also reported that higher frequency of cagA is in patients from high risk areas of gastric cancer than in those from low risk areas of gastric cancer. Some persons showed that cagA positive strains are associated with higher gastric cancer risk. But in our study, there was no significant difference in cagA status between the two areas. This provides further evidence on H pylori genotypes circulation in west and north China.
In conclusion, the present study showed the distinct distribution of H pylori virulence genotypes in two areas of north China with different gastric cancer risk. Further studies are required to determine the epidemiological and clinical importance of H pylori virulence-associated genotypes in different geographic areas in China.
| 1. | Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720-741. [PubMed] |
| 2. | Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137-5142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 564] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 3. | Marshall DG, Coleman DC, Sullivan DJ, Xia H, O'Moráin CA, Smyth CJ. Genomic DNA fingerprinting of clinical isolates of Helicobacter pylori using short oligonucleotide probes containing repetitive sequences. J Appl Bacteriol. 1996;81:509-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relation between clinical presentation, Helicobacter pylori density, interleukin 1beta and 8 production, and cagA status. Gut. 1999;45:804-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 87] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Atherton JC. H. pylori virulence factors. Br Med Bull. 1998;54:105-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 114] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111-2115. [PubMed] |
| 7. | van Doorn LJ, Figueiredo C, Sanna R, Blaser MJ, Quint WG. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J Clin Microbiol. 1999;37:2306-2311. [PubMed] |
| 8. | He Y, Hu PJ, He XX, Zeng ZR, Chen W, Peng XZ. The relationship between H pylori vacA genotypes and gastroenterology diseases in Guangzhou region. Weichang Bingxue. 2000;5:90-92. |
| 9. | Hou P, Xu G, Gong Y, Tu Z, Li Z, Ji X. The genotype of vac A gene of Helicobacter pylori (Hp) and its correlation with the gastroduodenal diseases associated with Hp. Zhonghua NeiKe ZaZhi. 1999;38:744-746. [PubMed] |
| 10. | Yuan Y, Zhang L. Comprehensive prevention for high-risk population from high-risk area of gastric cancer in China. Zhongguo Zhongliu. 2001;10:139-142. |
| 11. | Guo XL, Wang L, Wang L. The significant of serum HpcagA in the high-risk area of gastric cancer. World Chinese J Digestol. 2001;9:13-14. |
| 12. | Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839-2842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Tombola F, Del Giudice G, Papini E, Zoratti M. Blockers of VacA provide insights into the structure of the pore. Biophys J. 2000;79:863-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1108] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 15. | van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa JC, Carneiro F, Quint WG. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271-1276. [PubMed] |
| 16. | Cover TL, Tummuru MK, Cao P, Thompson SA, Blaser MJ. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566-10573. [PubMed] |
| 17. | Miehlke S, Yu J, Schuppler M, Frings C, Kirsch C, Negraszus N, Morgner A, Stolte M, Ehninger G, Bayerdörffer E. Helicobacter pylori vacA, iceA, and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am J Gastroenterol. 2001;96:1008-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Morales-Espinosa R, Castillo-Rojas G, Gonzalez-Valencia G, Ponce de León S, Cravioto A, Atherton JC, López-Vidal Y. Colonization of Mexican patients by multiple Helicobacter pylori strains with different vacA and cagA genotypes. J Clin Microbiol. 1999;37:3001-3004. [PubMed] |
| 19. | Sipponen P, Marshall BJ. Gastritis and gastric cancer. Western countries. Gastroenterol Clin North Am. 2000;29:579-592, v-vi. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Pan ZJ, Berg DE, van der Hulst RW, Su WW, Raudonikiene A, Xiao SD, Dankert J, Tytgat GN, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 96] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Peek RM, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, Miller GG. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531-544. [PubMed] |
| 22. | Dubois A, Berg DE, Incecik ET, Fiala N, Heman-Ackah LM, Del Valle J, Yang M, Wirth HP, Perez-Perez GI, Blaser MJ. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology. 1999;116:90-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | González-Valencia G, Atherton JC, Muñoz O, Dehesa M, la Garza AM, Torres J. Helicobacter pylori vacA and cagA genotypes in Mexican adults and children. J Infect Dis. 2000;182:1450-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |