Published online May 21, 2005. doi: 10.3748/wjg.v11.i19.2932
Revised: June 9, 2004
Accepted: August 5, 2004
Published online: May 21, 2005
AIM: To reveal the liver regeneration (LR) and its control as well as the occurrence of liver disease and to study the gene expression profiles of 551 genes after partial hepatectomy (PH) in regenerating rat livers.
METHODS: Five hundred and fifty-one expressed sequence tags screened by suppression subtractive hybridization were made into an in-house cDNA microarray, and the expressive genes and their expressive profiles in regenerating rat livers were analyzed by microarray and bioinformatics.
RESULTS: Three hundred of the analyzed 551 genes were up- or downregulated more than twofolds at one or more time points during LR. Most of the genes were up- or downregulated 2-5 folds, but the highest reached 90 folds of the control. One hundred and thirty-nine of them showed upregulation, 135 displayed downregulation, and up or down expression of 26 genes revealed a dependence on regenerating livers. The genes expressed in 24-h regenerating livers were much more than those in the others. Cluster analysis and generalization analysis showed that there were at least six distinct temporal patterns of gene expression in the regenerating livers, that is, genes were expressed in the immediate early phase, early phase, intermediate phase, early-late phase, late phase, terminal phase.
CONCLUSION: In LR, the number of down-regulated genes was almost similar to that of the upregulated genes; the successively altered genes were more than the rapidly transient genes. The temporal patterns of gene expression were similar 2 and 4 h, 12 and 16 h, 48 and 96 h, 72 and 144 h after PH. Microarray combined with suppressive subtractive hybridization can effectively identify the genes related to LR.
- Citation: Xu CS, Chang CF, Yuan JY, Li WQ, Han HP, Yang KJ, Zhao LF, Li YC, Zhang HY, Rahman S, Zhang JB. Expressed genes in regenerating rat liver after partial hepatectomy. World J Gastroenterol 2005; 11(19): 2932-2940
- URL: https://www.wjgnet.com/1007-9327/full/v11/i19/2932.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i19.2932
In the healthy adult rat liver, most of the hepatocytes lie in G0 phase, and their cell division index is very low (about one ten thousandth)[1-5]. However, metabolism of hepatocytes is quickly altered after partial hepatectomy (PH)[6-10]. Activation of hepatocytes in G0 phase occurs about 2 h after PH, and they progress to G1 phase about 6 h after PH. Then, the cells enter into S phase of cell cycle in 12 h. DNA synthesis occurs in the early 6 h (12-17 h) of S phase, and then DNA is synthesized 18-30 h after PH, which reaches a maximum at 24 h. The G2 phase of cell cycle lies in the subsequent 2-4 h (31-34 h after PH). After that, hepatocytes go on dividing, and the peak of cell division is at 36 h after PH. The next cycle of hepatocytes is in the following 36-66 h after PH[11,12]. The re-differentiation of liver cells and the re-building of regenerated livers are in 72-144 h after PH. Many experiments have confirmed that a cell cycle of hepatocytes lasts for about 30 h, but that of other cells distinguishes from them[13]. Briefly, cells in the residual liver would be activated to proliferate, re-differentiate and rebuild their structure and function after PH. In different phases of liver regeneration (LR), the physiological and biochemical actions of different kinds of cells of the liver are different. The categories and amounts of the expressed genes in them are various[14,15]. To learn the molecular mechanism of LR, it is essential to highlight how many genes are related to it. Therefore, this paper reports that 300 genes have been successfully identified to correlate with LR by combing microarray in combination with suppression subtractive hybridization.
Healthy SD rats weighing 200±20 g were obtained from the Experimental Animal Center of Henan Normal University. Following the method of Higgins and Anderson[16], 70% of the rat liver was removed under sterile conditions.
The regenerating livers of four rats (male:female = 1:1) were taken 2, 4, 8, 12, 16, 24, 36, 48, 72, 96 and 144 h after PH. The livers were rinsed in cold PBS and immersed in a -80 °C refrigerator for RNA extraction. Total RNA was isolated from frozen livers according to the manual of TRIzol kit of Invitrogen. In brief, 50-100 mg liver was homogenized in 1 mL TRIzol reagent containing phenol and guanidinium isothiocyanate/cationic detergent, followed by phenol-chloroform extraction and isopropyl alcohol precipitation. The quantity and integrity of total RNA were examined by an ultraviolet spectrometer and denaturing formaldehyde agarose electrophoresis by ethidium bromide staining.
cDNA subtracted libraries were generated from total RNA by PCR-SelectTM cDNA subtraction kit (Clontech) following the manufacturer’s instructions. Briefly, total RNA was transcripted into double cDNA strands and digested with restriction enzymes, followed by subtracted hybridization with drivers and testers. Finally, differential expression sequence tags were performed to construct subtracted cDNA libraries with suppression PCR.
Base sequence assay of ESTs was carried out according to the current protocols in molecular biology. All sequences were determined on both strands. Comparison analysis of the selected sequences was conducted with the DNAman and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) GenBank database[17].
cDNA fragments amplified by PCR with nested PCR primer1 and primer2 and purified by NaAc/isopropyl alcohol were spotted onto glass slides (BioStar) with the help of the ProSys-5510A spotting machine. Then the gene chips were ready by hydrating and blocking and drying. A total of 1 152 elements (double spot chip) including 50 control system genes (8 negative control, 12 void control, 30 internal control) and 551 target genes to be studied comprised 8 submatrixes (12*12) occupying 9 mm×18 mm (BioStar). Then the gene chips were ready by hydrating, blocking and drying.
RNA prepared from rat livers before PH was ready for a reference for all cDNA microarray analyses. Total denatured RNA was reverse transcribed with cy3-conjugated dCTP (control group) and cy5-conjugated dCTP (test group) (Amersham-Pharmacia Biotech) using MMLV reverse transcriptase (Promega) with oligo(dT) primers. After bath incubation for 2 h, labeled buffers I and II were subsequently added to the reaction. The control group and test group were mixed symmetrically and stored avoiding light for application[17].
Glass slices were prehybridized at 42 °C for 5-6 h in hybridization buffer containing freshly cooked salmon sperm DNA. The labeled denatured probes were hybridized against cDNA microarray and incubated overnight (16-18 h) at 42 °C. The slices were then washed twice with 2×SSC containing 0.5% SDS for 5 min at room temperature, then with 0.2×SSC containing 0.5% SDS at 60 °C for 10 min, and finally with 0.2×SSC at 60 °C for 10 min. The slices were exposed to a photographer. Hybridized images were scanned by a fluorescence laser-scanning device, GenePix4000A. At least, two hybridizations were performed at each time point. In addition, a semiquantitative inspection of the hybridization results was performed for green signals (downregulation), yellow signals (no obvious regulation), and red signals (upregulation)[17].
cy3 and cy5 signal intensities were quantified by GenePixPro 3.0 software. Subsequently, we normalized the obtained numerical data with classical linear regression techniques. In brief, quantified cy3 and cy5 signal intensities were obtained when foreground signal intensities were deducted by background signal intensities and cy5 signal intensities were replaced by 200 when they were <200. When Ri (Ri = cy5/cy3) was between 0.1 and 10, Ri was taken as logarithms to generate Ri’ [log(Ri)] and ND was taken by EXP (R) (averaged Ri’). The modified cy3* was generated by ND multiplying cy3 and was replaced by 200 when it was <200. The ratio was expressed by cy5/cy3*. Therefore, we selected the genes, whose ratio was more than 2 or less than 0.5 representing a twofold difference in expression level. To analyze the selected gene expression data, we performed GeneMaths cluster analysis and hierarchical clustering to appraise the number of groups. Whole analyses were executed using Microsoft Excel and GeneSpring[11,17].
The analysis of gene expression spectrum showed that, of the 551 elements made in microarray, the expression intensity of 300 genes increased or decreased at least more than twofolds at one time point after PH. Their sequence analysis showed that 152 were unreported genes, and 133 genes were significantly reported. Of which 49 were upregulated in regenerating rat livers, 74 were downregulated, and 10 were either up- or downregulated in different phases of LR. Following biochemical actions, the 133 elements were categorized into 24 groups (Table 1).
| No. | Gene description | Fold difference |
| Stress response | ||
| 1 | Alpha-1 major acute phase protein | 6.2 |
| 2 | Acute-phase protein alpha-1-inhibitor 3 | 0.2 |
| 3 | Clusterin (Clu) | 0.3 |
| 4 | C-reactive protein, petaxin related | 2.2 |
| Glycometabolism | ||
| 5 | Isocitrate dehydrogenase 1 (Idh1) | 0.3 |
| 6 | Glycerol 3-phosphate dehydrogenase (Gpd3) | 0.4 |
| 7 | Aldolase B | 0.3 |
| 8 | Enolase 1 | 4 |
| 9 | Small subunit of RuBPCase | 3.3, 0.1 |
| Fatty and stearoyl metabolism | ||
| 10 | Acyl-CoA desaturase | 0.4 |
| 11 | 3-alpha-hydroxysteroid dehydrogenase | 0.2 |
| 12 | Bile acid CoA ligase | 2.3 |
| 13 | Methylmalonate semialdehyde dehydrogenase | 0.3 |
| 14 | Prostaglandin D2 synthase 2 (Ptgds2) | 3 |
| 15 | Cytochrome P450 cholesterol 7-alpha-hydroxylase (P450 VII) | 2.3, 0.5 |
| 16 | Malonyl-CoA decarboxylase | 0.3 |
| 17 | NAD(P)-dependent steroid dehydrogenase | 0.4 |
| Oxidation and reduction response | ||
| 18 | Acyl-coA oxidase (RATACOA1) | 2.7, 0.4 |
| 19 | Cytochrome b5 (Cyb5) | 0.2 |
| 20 | Flavin-containing monooxygenase 1 (Fmo1) | 0.1 |
| 21 | Paraoxonase 1 (Pon1) | 0.2 |
| 22 | Selenium-dependent glutathione peroxidase | 0.4 |
| 23 | Peroxisomal sarcosine oxidase (PSO) | 0.3 |
| 24 | P450 arachidonic acid epoxygenase (cyp 2C23) | 0.2 |
| 25 | Cytochrome P450 | 0.2 |
| 26 | Cytochrome P450 15-beta (Cyp2c12) | 0.2 |
| 27 | Cytochrome P450 2E1 | 0.1 |
| 28 | Cytochrome P450, 2c39 (Cyp2c39) | 0.1 |
| 29 | Cytochrome P450, 7a1 (Cyp7a1) | 2 |
| 30 | Cytochrome P450 (PNCN inducible, Cyp3A1) | 0.2 |
| 31 | Cytochrome b | 0.5 |
| Regulation proteins | ||
| 32 | G-protein, beta polypeptide 2 (Gnb2l1) | 2.4 |
| 33 | Interferon-induced protein with tetratricopeptide repeats 3 | 0.2 |
| 34 | Protein RAKb | 0.2 |
| Glycoproteins | ||
| 35 | Selenoprotein P (Sepp1) | 0.3 |
| 36 | Myelin-associated glycoprotein (L-MAG) | 7 |
| 37 | Alpha-1-B glycoprotein (A1bg) | 0.1 |
| 38 | Histidine-rich glycoprotein (Hrg) | 0.1 |
| 39 | Mannosylalpha-1, 6-glycoprotein beta-1, 2-N-acetylglucosaminyltransferase (Mgat2) | 2 |
| 40 | UDP-glucuronosyltransferase 2B3 (Udpgt) | 0.3 |
| 41 | Fibrinogen gamma polypeptide (Fgg) | 5.1 |
| 42 | TRAM1 | 3.3, 0.4 |
| Lipid-proteins | ||
| 43 | Apolipoprotein C-II | 0.3 |
| 44 | Apolipoprotein M (Apom) | 0.4 |
| 45 | Fatty acid binding protein 1 (Fabp1) | 0.3 |
| 46 | Retinol-binding protein(PRBP) | 0.4 |
| 47 | Solute carrier family 20 (phosphate transporter) member 1 (Slc20a1) | 0.4 |
| 48 | Transthyretin-related protein (TTN) | 0.3 |
| Nucleolar proteins | ||
| 49 | P53 | 2 |
| 50 | Damage-specific DNA binding protein 1 (Ddb1) | 0.3 |
| 51 | Nucleolar protein family A, member 2 | 3.9 |
| 52 | Nuclear protein 1 (Nupr1) | 5.1 |
| 53 | AP1 gamma subunit binding protein 1 | 2.3 |
| Receptors | ||
| 54 | Interleukin 1 receptor, type I (Il1r1) | 7.9 |
| 55 | Golgi SNAP receptor complex member 1 (Gosr1) | 0.4 |
| 56 | Nuclear receptor subfamily 0, member 2 (Nr0b2) | 0.2 |
| 57 | Lysosomal-associated protein transmembrane 4A | 0.5 |
| 58 | ATP-binding cassette, sub-family B | 0.4 |
| 59 | ATP-binding cassette, sub-family C | 0.2 |
| Factors | ||
| 60 | Eukaryotic translation initiation factor 4A1 | 3.8 |
| 61 | Eukaryotic release factor 3 | 2 |
| 62 | Insulin-like growth factor I | 0.5 |
| 63 | Early growth response factor 1 (Egr1) | 3.6 |
| 64 | Neuropeptide Y (Npy) | 18.2 |
| 65 | NF-E2-related factor 2 (Nfe2l2) | 0.4 |
| 66 | Pre-B-cell colony-enhancing factor (Pbef) | 3.5 |
| 67 | Amphoterin | 0.3 |
| 68 | Angiogenin | 0.2 |
| 69 | Angiopoietin-like 3 | 0.2 |
| 70 | Ring1 and YY1 binding protein | 0.5 |
| 71 | Chromatin remodeling factor | 2 |
| Hemoglobins | ||
| 72 | Hemoglobin beta chain (Hbb) | 0.3 |
| Immunological proteins | ||
| 73 | Immunoglobulin C kappa | 0.2 |
| 74 | Achaete-scute complex homolog-like 1 (Ascl1) | 0.4 |
| 75 | Complement component 1 | 0.3 |
| 76 | Complement component 5 | 8.8 |
| 77 | JE/MCP-1 | 4 |
| 78 | Class III Fc-gamma receptor | 3.5 |
| Chaperonins | ||
| 79 | 70 ku heat shock protein (Hspa5) | 2.8, 0.5 |
| 80 | DnaJ (Hsp40) | 2.5 |
| Cytoskelets | ||
| 81 | Actin gamma | 4.7 |
| 82 | Karyopherin (importin) alpha 2 | 0.4 |
| 83 | Clathrin, heavy polypeptide (Cltc) | 3.3 |
| Ribosomal proteins | ||
| 84 | Ribosomal protein L28 (Rpl28) | 2.2 |
| 85 | Ribosomal protein L41 (Rpl41) | 2.1 |
| 86 | Ribosomal protein S19 | 2.5 |
| Marker proteins | ||
| 87 | Probable surface antigen protein | 2.1, 0.4 |
| 88 | Surfeit 1 (Surf1) | 2.5 |
| 89 | Alpha globin | 0.3 |
| 90 | Pregnancy-zone protein (Pzp) | 2.4 |
| 91 | Adipose differentiation-related protein | 7.3 |
| 92 | Subchromosomal-transferable fragment 4 | 2.3, 0.3 |
| 93 | Cocoa protein | 4.9 |
| 94 | Amyloid P-component (Sap) | 0.4 |
| 95 | Amyloid alpha-5 protein | 0.2 |
| Amino acid enzymes | ||
| 96 | Arginase 1 (Arg1) | 0.4 |
| 97 | 2-hydroxyphytanoyl-CoA lyase (Hpcl2) | 90.5 |
| 98 | Cytosolic aspartate aminotransferase | 5.3 |
| Nuclearase | ||
| 99 | Mitochondrial adenine nuleotidase | 2 |
| 100 | Rnase A family 4 | 0.2 |
| 101 | Rnase H | 0.4 |
| Proteolytic enzymes | ||
| 102 | Caspase 1 (Casp1) | 2.6 |
| 103 | Cathepsin C (Ctsc) | 0.4 |
| 104 | Cathepsin D (Ctsd) | 0.3 |
| 105 | Neutrophil collagenase (Mmp8) | 0.2 |
| 106 | Coagulation factor 2 (F2) | 0.2 |
| Proteinase inhibitors | ||
| 107 | Alpha-1 microglobulin/bikunin (Ambp) | 2.1, 0.2 |
| 108 | Alpha-2-macroglobulin (A2m) | 21.3 |
| 109 | Alpha-trypsin inhibitor, heavy chain 4 | 0.2 |
| 110 | Leuserpin-2 (Serpind1) | 0.2 |
| 111 | Serine protease inhibitor 1 | 5 |
| Phopshorylases | ||
| 112 | Thymidylate kinase (dTMP kinase) | 3 |
| 113 | Mss4 protein | 2.1 |
| 114 | CDK110 | 0.5 |
| Phosphatases | ||
| 115 | Carbamyl phosphate synthetase I | 2.9 |
| 116 | Phosphodiesterase 1(Enpp1) | 6.6 |
| 117 | Phosphatidylserine-specific phospholipase A1 | 2.8 |
| 118 | Protein phosphatase 1 (GL-subunit) | 0.2 |
| 119 | UTP-glucose-1-phosphatase | 0.4 |
| Synthase | ||
| 120 | H+-ATP synthase alpha subunit (Atp5a1) | 2.4, 0.4 |
| 121 | Bifunctional aminoacyl-tRNA synthetase | 0.4 |
| 122 | ATP synthase subunit 8 | 2.2 |
| 123 | Glutamyl-prolyl-tRNA synthetase (Eprs) | 4.5 |
| 124 | Fatty acid elongase 1 (rELO1) | 0.2 |
| 125 | RNA cyclase | 0.2 |
| Transferases | ||
| 126 | Carnitine O-octanoyltransferase (Crot) | 0.3 |
| 127 | Glutathione S-transferase, alpha 1 (Gsta1) | 0.1 |
| 128 | Glutathione S-transferase, type 3 (Gstm3) | 2.2, 0.3 |
| 129 | Microsomal glutathione S-transferase 1 (Mgst1) | 0.4 |
| 130 | Sulfotransferase K2 | 0.3 |
| 131 | Sialyltransferase 1 (Siat1) | 2.6 |
| 132 | UDP-glucuronosyltransferase member | 50.3 |
| (Ugt2b5) | 0.5 | |
| 133 | Protein disulfide isomerase-related protein |
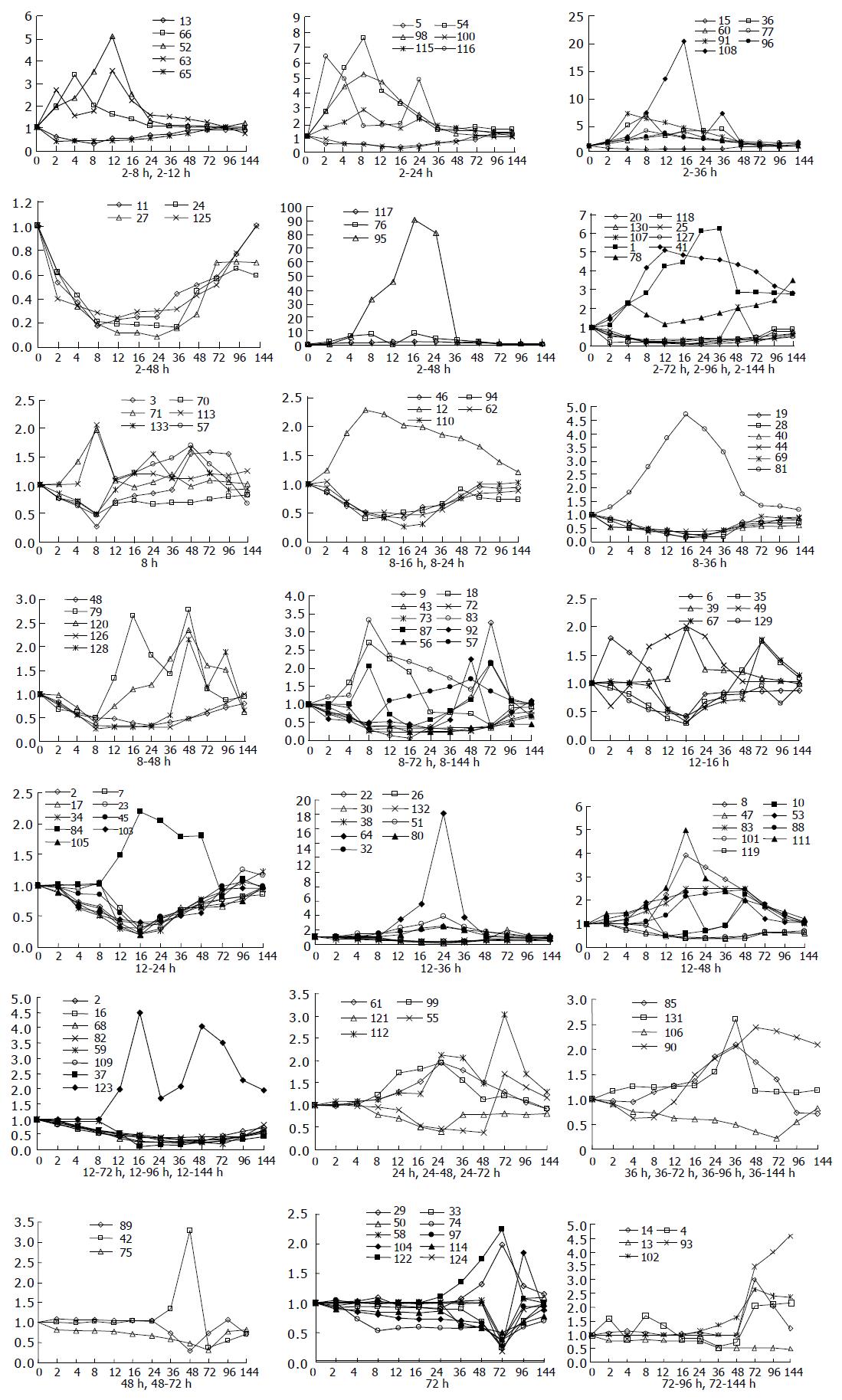
Analysis of their expression in the regenerating livers at different time points after PH showed that, in the priming phase of LR, the expression of 34 genes was rapidly altered within 6 h and then recovered gradually to the control levels in 48-72 h after PH. In the cell cycle progression phase of 8-36 h after PH, the expression of 86 genes was altered markedly. In the terminal phase of LR over 72 h after PH, the expression of 13 genes was altered distinctly (Figure 1).
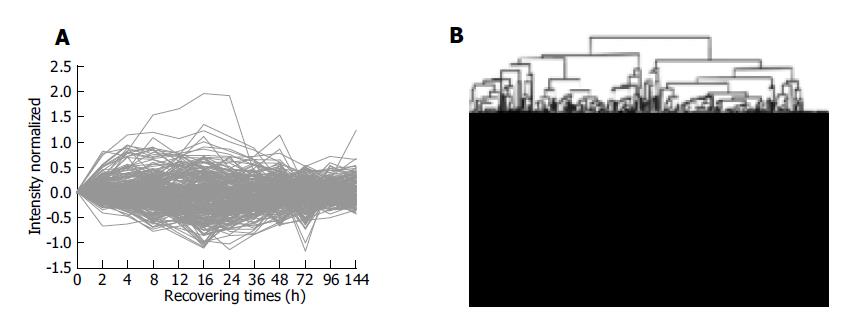
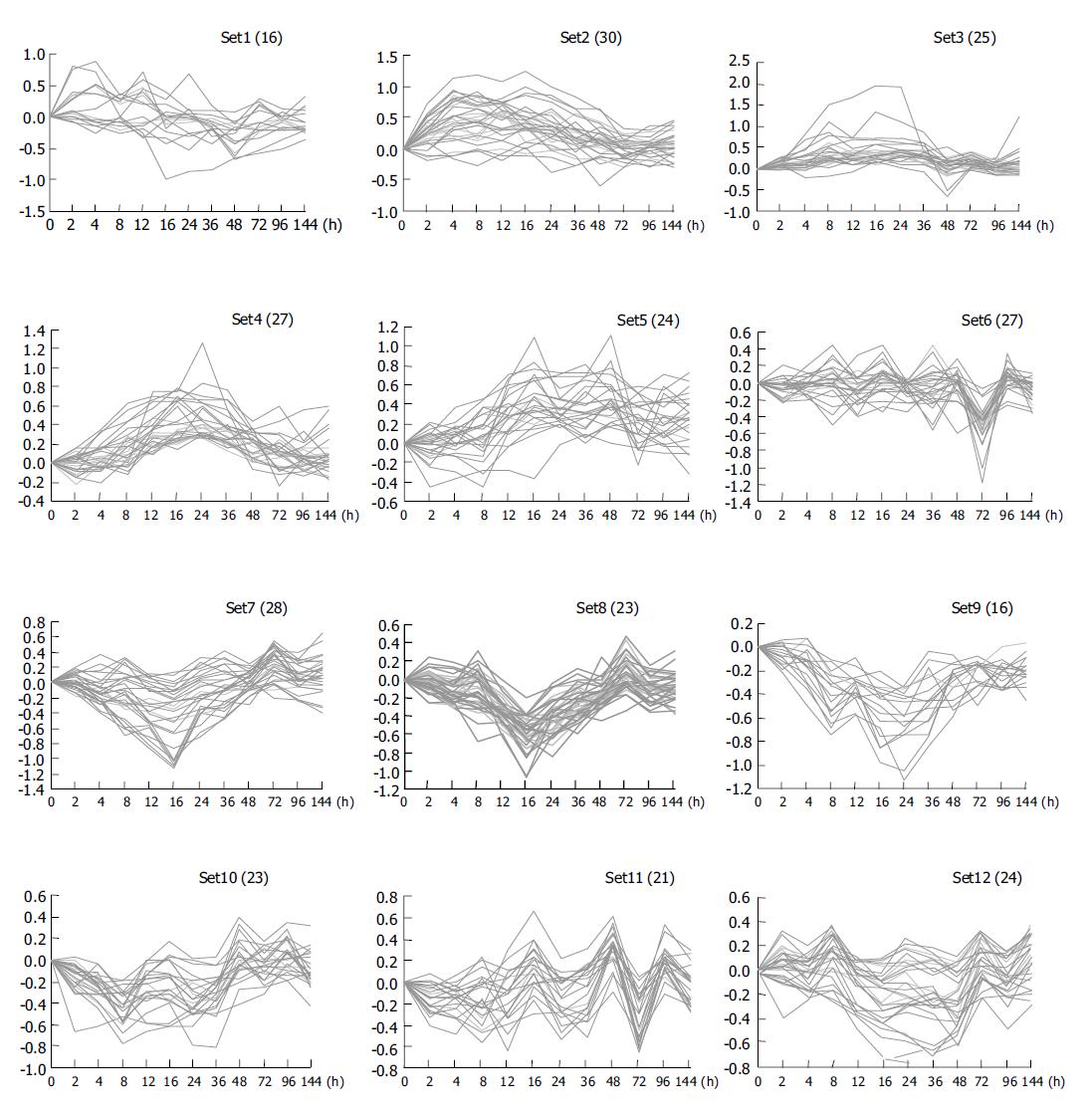
Clustering analysis of genes expressed at 12 time points after PH showed that the 300 elements altered at least more than twofolds in density at one time point (Figure 2A), and that the most similar patterns of gene expression were located next to each other and placed in a major branch of the dendrogram. Twelve and sixteen hours 48 and 96 h, 72 and 144 h patterns were clustered as separate groups in major branches, indicating that these time points shared common expression profiles of genes (Figure 2B). Thirty-four genes were induced to express in 2-4 h and reached a maximum in 8-24 h, but a smaller peak of gene expression appeared at 72 h after PH. To facilitate the visualization and interpretation of the gene expression program in these data, we used the method of GeneMaths to order genes of similarities in their expression patterns and displayed them in a compact graphical format (Figure 3). Based on the expressed characteristics of genes in LR and following the review of Michalopoulos and Defrances, the selected elements were categorized into six distinct temporal patterns of expression: the genes of rapid induction which were expressed in the immediate early phase of 2-6 h after PH, the genes of early induction in the early phase of 8-16 h, the genes of middle induction in the intermediate phase of 16-24 h, the genes of early-late induction in the early-late phase of 24-36 h, the genes of late induction in the late phase of 36-72 h, and the genes of consistent repression in the terminal phase of 72-144 h.
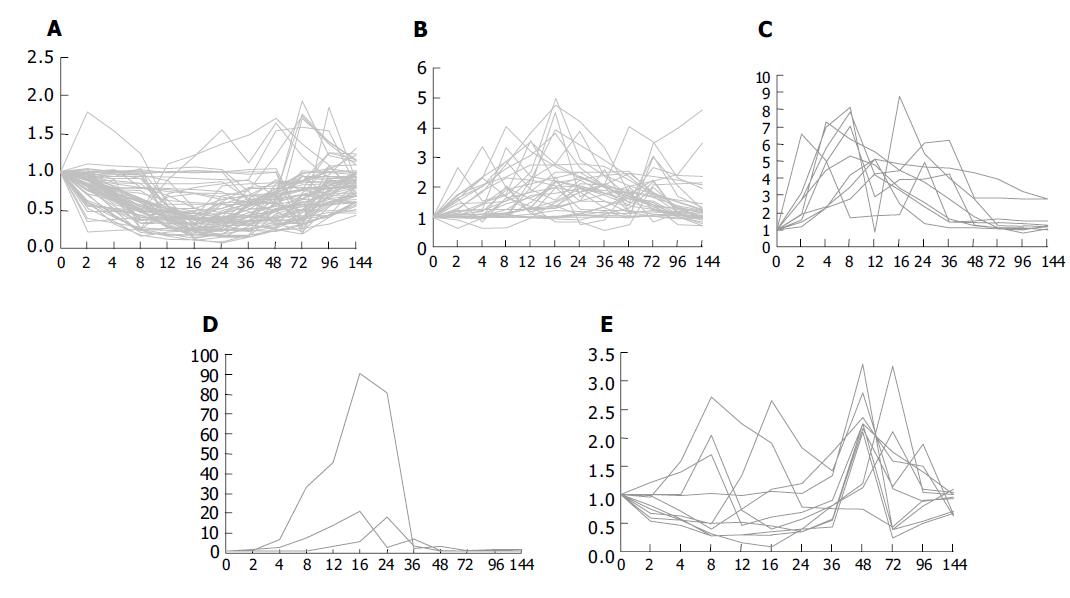
It was found that expression of the 133 genes in the regenerating livers was quite different. Seventy-four of them decreased 2-10 folds, 49 of them were increased to 2-5 folds, 9 genes to 5-10 folds, 3 genes to over 10 folds, and the maximum to over 90 folds (Figure 4).
In the immediate early phase of LR (2-6 h after PH), liver damage occurred in inflammation response, hepatocytes in G0 phase were activated, and then progressed to G1 phase. Thirty-four genes, except the previous reported genes, were consistently and rapidly induced to express, 19 of them were upregulated and 15 downregulated. Among the genes, NF-E2-related factor 2 (Nfe2l2), isocitrate dehydrogenase, JE/MCP-1, and complement component 5 were advantageous to block oxidative injuries and to prevent the occurrence of inflammation of the residual liver after PH. Pre-B-cell colony-enhancing factor (PBEF), nuclear protein 1 (Nupr1), early growth response factor 1 (Egr1) were upexpressed, which probably play an important role in activating hepatocytes to start DNA synthesis[18]. The expression of isozymes of aspartate aminotransferase and phosphodiesterase 1 (Enpp1) was induced, which supply purine and adenosine for DNA synthesis[19]. The proteins of providing energy for LR such as adipose differentiation-related protein and phosphatidylserine-specific phospholipase A1 and proteins associated with inflammation response and cell apoptosis such as α-1 major acute phase protein, fibrinogen gamma polypeptide (Fgg), kininogen and class III Fc-gamma receptor were also included, which play important parts in preventing the occurrence of inflammation and the startup of LR in the immediate early phase.
In the early phase of LR (8-16 h after PH), livers damaged stress response and hepatocytes were prepared to synthesize DNA. The expression of 31 genes was altered at 8 h after PH. Among them, 11 were upregulated and 20 downregulated, of which, 5 genes reached expression peak at 8 h after PH. Myelin-associated glycoprotein (MAG) was induced, which is important for nerve system regeneration in LR[20]. The stress-inducible 70-ku protein (Hsp70) and protein disulfide isomerase-related protein were expressed, which promote protein folding correctly and degradation of harmful proteins in LR. Serum amyloid P component and clusterin were repressed in 8-16 h after PH, which can decrease accumulation of false or toxic proteins in neurocytes, and are useful in protecting neonatal livers[21]. Plasma retinol-binding protein, bile acid CoA ligase (BAL), insulin-like growth factor-I (IGF-I), ApoM, angiopoietin-like protein 3 (Angptl3) and lysosomal-associated protein transmembrane-4 beta (LAPTM4B) were increased as well in this phase. Plasma retinol-binding protein could transport retinol and vitamin A, whose increase in 8-16 h after PH showed that retinol and vitamin A were essential to hormone synthesis in LR[22]. BAL could facilitate adsorption of amino acids by catalyzing conjugation of bile acids with amino acids[23]. IGF-I was found to be involved in the liver and brain growth and development of embryos[24]. Its downregulation in 8-24 h after PH indicated that cell cycle was in the phase of DNA synthesis. The reduction of ApoM in 8-24 h after PH was supposed to promote lipolysis and to provide energy for LR. Angptl3 is a hepatic secretary factor, which could activate lipolysis in adipocytes by response to the liver X receptor[25]. LAPTM4B, a novel gene was upregulated in hepatic carcinoma, whose N-terminus is essential for the survival of cells[26]. The downregulation of LAPTM4B may be related to refraining cell necrosis in LR.
In the intermediate phase of LR (16-24 h after PH), hepatocytes synthesized DNA. It was confirmed that 41 genes were expressed at 12 h after PH. The increase of mitochondrial glycerol 3-phosphate dehydrogenase (mGPDH) at 12 h was assumed to provide enough ATP for DNA synthesis. NAD(P)H steroid dehydrogenase (Nsdhl), hepatic cytochrome P450 cholesterol 7 alpha-hydroxylase (CYP7) and ATP-binding cassette were induced in this time, which may play a role in the conversion of lanosterol into cholesterol[27,28]. L28 is a component of 70S ribosome, whose upregulation in 12-24 h after PH was supposed to help 70S ribosome conformation and to accelerate protein synthesis. Activator protein 1 (AP1)-like elements and P53 were increased, which probably prevent injured liver from apoptosis and necrosis[29]. The proteins associated with protein fold such as cathepsin C (Ctsc), dipeptidyl aminopeptidase I (Dpap1), Hsp40 and hsp70 (p73/p72) were also up expressed, which are essential to form the correct structure of proteins.
In the early-late phase of LR (24-36 h after PH), hepatocytes must complete all the activations of later S phase, G2 phase and M phase. It was checked that seven genes were started to express at 24 h after PH. Five of them were upregulated and two were downregulated, of which, the restrain of bifunctional aminoacyl-tRNA synthetase was up expressed at 24 h after PH, which may play a pivotal role in separating the subunits of the eukaryotic tRNA synthetase complex in LR. Furthermore, eukaryotic release factor 3 incorporated in a complete scheme for translation termination of proteins, whose induction may be advantageous to release essential proteins to exert functions. The mRNA level of sialyltransferase was increased at 36 h, suggesting that liver releases large amounts of sialyltransferase to mediate recovery of liver functions after PH.
In the late phase of LR (24-36 h after PH), hepatocytes went through the second cell cycle, and other cells began to divide. It was found that seven genes were induced to express at 36 h after PH. Of them, coagulation factor 2 inhibitor was upregulated, which facilitates blood circulation in the regenerating liver. The pregnancy-zone protein (Pzp) was induced, which accelerates hepatocyte division and mediate cell differentiation in LR. Complement component 1 was increased in 48-72 h after PH, which protects against the outer intruders in the injured liver.
In the terminal phase of LR (72-144 h after PH), structure and function of the regenerating liver were recovered. It was found that 15 genes were induced in 48-144 h after PH. Of them, galactose-specific genes such as membrane-bound C-reactive protein were increased, demonstrating that galactose is necessary in terminal phase of LR. Cathepsin D was decreased, suggesting that cell migration was suppressed[30]. CDK110 was upregulated at 72 h after PH, which mediates cell differentiation in LR. Flavanol cocoa was continuously induced in LR, which can prevent the regenerated liver from early alcohol-induced liver injury[31].
In summary, we have screened 576 genes from subtracted cDNA libraries and made an in-house cDNA microarray. These genes were highly and specifically expressed in the liver. Using the chips, we performed a large-scale analysis of gene expression in LR and found that the expressions of 133 reported and 167 unreported genes were altered more than twofolds at one or more time points. Cluster analysis showed gene expression patterns at 2 and 4 h, 12 and 16 h, 48 and 96 h as well as 72 and 144 h after PH had strong correlations respectively. The 133 reported genes could be categorized into six distinct temporal patterns of induction and were involved in 24 groups of proteins. Their actions in LR were discussed. However, to elucidate the mechanism of LR, further research is needed.
The authors thank BioStar for providing microarray.
| 1. | Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2649] [Cited by in RCA: 2468] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 2. | Fausto N. Liver regeneration. J Hepatol. 2000;32:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 912] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 3. | Zimmermann A. Liver regeneration: the emergence of new pathways. Med Sci Monit. 2002;8:RA53-RA63. [PubMed] |
| 4. | Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413-427. [PubMed] |
| 5. | Nagy P, Bisgaard HC, Schnur J, Thorgeirsson SS. Studies on hepatic gene expression in different liver regenerative models. Biochem Biophys Res Commun. 2000;272:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Gressner AM. Cytokines and cellular crosstalk involved in the activation of fat-storing cells. J Hepatol. 1995;22:28-36. [PubMed] |
| 7. | Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21:1443-1449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 236] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 8. | FitzGerald MJ, Webber EM, Donovan JR, Fausto N. Rapid DNA binding by nuclear factor kappa B in hepatocytes at the start of liver regeneration. Cell Growth Differ. 1995;6:417-427. [PubMed] |
| 9. | Hsu JC, Laz T, Mohn KL, Taub R. Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc Natl Acad Sci U S A. 1991;88:3511-3515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Enami Y, Kato H, Murakami M, Fujioka T, Aoki T, Niiya T, Murai N, Ohtsuka K, Kusano M. Anti-transforming growth factor-beta1 antibody transiently enhances DNA synthesis during liver regeneration after partial hepatectomy in rats. J Hepatobiliary Pancreat Surg. 2001;8:250-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Sato Y, Igarashi Y, Hakamata Y, Murakami T, Kaneko T, Takahashi M, Seo N, Kobayashi E. Establishment of Alb-DsRed2 transgenic rat for liver regeneration research. Biochem Biophys Res Commun. 2003;311:478-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Fukuhara Y, Hirasawa A, Li XK, Kawasaki M, Fujino M, Funeshima N, Katsuma S, Shiojima S, Yamada M, Okuyama T. Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 2003;38:784-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Xu CS, Lu AL, Xia M, Li XY, Li YH, Zhao XY. The effect of heat shock before rat partial hepatectomy on HSC70/HSP68 expression and phosphatase activities. ShiYan ShengWu XueBao. 2000;33:1-11. [PubMed] |
| 14. | Jensen SA. Liver gene regulation in rats following both 70 or 90% hepatectomy and endotoxin treatment. J Gastroenterol Hepatol. 2001;16:525-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci U S A. 2002;99:11181-11186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Higgins GM, Anderson RM. Experimental pathology of the liver I. Restoration of the liver of white rat following partial surgical removal. Arch Pathol. 1931;12:186-202. |
| 17. | Yang GP, Ross DT, Kuang WW, Brown PO, Weigel RJ. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res. 1999;27:1517-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 170] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 18. | Ognjanovic S, Bao S, Yamamoto SY, Garibay-Tupas J, Samal B, Bryant-Greenwood GD. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J Mol Endocrinol. 2001;26:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 187] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Kamimoto Y, Horiuchi S, Tanase S, Morino Y. Plasma clearance of intravenously injected aspartate aminotransferase isozymes: evidence for preferential uptake by sinusoidal liver cells. Hepatology. 1985;5:367-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Ito H, Ishida H, Collins BE, Fromholt SE, Schnaar RL, Kiso M. Systematic synthesis and MAG-binding activity of novel sulfated GM1b analogues as mimics of Chol-1 (alpha-series) gangliosides: highly active ligands for neural siglecs. Carbohydr Res. 2003;338:1621-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Hind CR, Collins PM, Pepys MB. Calcium-dependent aggregation of human serum amyloid P component. Inhibition by the cyclic 4,6-pyruvate acetal of galactose. Biochim Biophys Acta. 1984;802:148-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Soprano DR, Pickett CB, Smith JE, Goodman DS. Biosynthesis of plasma retinol-binding protein in liver as a larger molecular weight precursor. J Biol Chem. 1981;256:8256-8258. [PubMed] |
| 23. | Falany CN, Xie X, Wheeler JB, Wang J, Smith M, He D, Barnes S. Molecular cloning and expression of rat liver bile acid CoA ligase. J Lipid Res. 2002;43:2062-2071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Santos A, Yusta B, Fernández-Moreno MD, Blázquez E. Expression of insulin-like growth factor-I (IGF-I) receptor gene in rat brain and liver during development and in regenerating adult rat liver. Mol Cell Endocrinol. 1994;101:85-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Shimamura M, Matsuda M, Kobayashi S, Ando Y, Ono M, Koishi R, Furukawa H, Makishima M, Shimomura I. Angiopoietin-like protein 3, a hepatic secretory factor, activates lipolysis in adipocytes. Biochem Biophys Res Commun. 2003;301:604-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Shao GZ, Zhou RL, Zhang QY, Zhang Y, Liu JJ, Rui JA, Wei X, Ye DX. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene. 2003;22:5060-5069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Ohashi M, Mizushima N, Kabeya Y, Yoshimori T. Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J Biol Chem. 2003;278:36819-36829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Hoekstra M, Kruijt JK, Van Eck M, Van Berkel TJ. Specific gene expression of ATP-binding cassette transporters and nuclear hormone receptors in rat liver parenchymal, endothelial, and Kupffer cells. J Biol Chem. 2003;278:25448-25453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 148] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 29. | Miró F, Lelong JC, Pancetti F, Roher N, Duthu A, Plana M, Bourdon JC, Bachs O, May E, Itarte E. Tumour suppressor protein p53 released by nuclease digestion increases at the onset of rat liver regeneration. J Hepatol. 1999;31:306-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 30. | Bond JS, Aronson NN. Endocytosis and degradation of native, cathepsin D-degraded, and glutathione-inactivated aldolase by perfused rat liver. Arch Biochem Biophys. 1983;227:367-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 31. | McKim SE, Konno A, Gäbele E, Uesugi T, Froh M, Sies H, Thurman RG, Arteel GE. Cocoa extract protects against early alcohol-induced liver injury in the rat. Arch Biochem Biophys. 2002;406:40-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |