Published online May 1, 2004. doi: 10.3748/wjg.v10.i9.1301
Revised: March 20, 2003
Accepted: April 11, 2003
Published online: May 1, 2004
AIM: To construct a differentially-expressed gene subtracted cDNA library from two colorectal carcinoma (CRC) cell lines with different metastatic phenotypes by suppression subtractive hybridization.
METHODS: Two cell lines of human CRC from the same patient were used. SW620 cell line showing highly metastatic potential was regarded as tester in the forward subtractive hybridization, while SW480 cell line with lowly metastatic potential was treated as tester in the reverse hybridization. Suppression subtractive hybridization (SSH) was employed to obtain cDNA fragments of differentially expressed genes for the metastasis of CRC. These fragments were ligated with T vectors, screened through the blue-white screening system to establish cDNA library.
RESULTS: After the blue-white screening, 235 white clones were picked out from the positive-going hybridization and 232 from the reverse. PCR results showed that 200-700 bp inserts were seen in 98% and 91% clones from the forward and reverse hybridizations, respectively.
CONCLUSIONS: A subtractive cDNA library of differentially expressed genes specific for metastasis of CRC can be constructed with SSH and T/A cloning techniques.
- Citation: Liang L, Ding YQ, Li X, Yang GZ, Xiao J, Lu LC, Zhang JH. Construction of a metastasis-associated gene subtracted cDNA library of human colorectal carcinoma by suppression subtraction hybridization. World J Gastroenterol 2004; 10(9): 1301-1305
- URL: https://www.wjgnet.com/1007-9327/full/v10/i9/1301.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i9.1301
CRC is one of the most common malignant tumors in the world and its metastasis is the major cause of mortality in patients with colorectal carcinoma. More than hundreds of genes have been reported to be involved in the regulations of metastasis in colorectal carcinoma. However, they are still not sufficient for fully explaining the complexity and diversity of metastasis. Besides, current investigations of these genes that mostly focused on the expression analysis of one or several genes make it difficult to understand the genes’ interactions and find the new genes. Cloning and identification of metastasis-associated genes have been hypothesized to be beneficial to the elucidation of the molecular mechanisms underlying the metastasis and finding gene targets for metastatic forecast, therapy and prognosis of CRC patients. In our study, SSH and bacterial culture PCR screening were used to construct a differentially-expressed gene subtracted cDNA library specific for metastasis of human CRC with the hope to screen new metastasis associated genes.
Highly metastatic cell line SW620 and lowly metastatic cell line SW480 from human colorectal carcinoma were purchased from ATCC with the number of CCL-227 and CCL-228 respectively. Both of the paired cell lines were from one colonic adenocarcinoma patient and had the same genetic backgrounds. Cell lines were routinely cultured with DMEM supplemented with 10% bovine serum under the atmosphere containing 50 mL/L CO2 at 37 °C.
cDNA synthesis primer (5’-TTTTGTACAAGCTT30N1N-3’) was used for the first-strand cDNA synthesis. Adaptor1 (5’-CTAATACGACTCACTATAGGGCTCGAGCGGCCGCCCGGG CAGGTACCTGCCCGG-3’) and adaptor2R (5’CTAATACGAC TCACTATAGGGCAGCGTGGTCGCGGCCGAGGTACCTCGGC CG-3’) have the same sequence of the 5’ region and the different palindromic structures in the 3’ end. Only the differentially expressed fragments digested by RsaI restriction enzyme could be ligated with two adaptors. Primary PCR was performed to amplify these fragments with primer1 (5’-CTAATACGACTCAC TATAGGGC-3’) that corresponds to the outer common sequence of the adaptors. After that, nested primer1 (5’-TCGAGCGGCC GCCCGGGCAGGT-3’) and nested primer2R (5’-AGCGTGG TCGCGGCCGAGGT-3’) designed from the inner sequences of the adaptors respectively were used for the further enrichment of these fragments in secondary PCR. The underlines in the sequences indicated the sites of RsaI restriction enzyme. The above primers and adaptors were provided by PCR-Select TMcDNA Subtraction Kit (Clontech Laboratories Inc, United States).
Total mRNA was isolated from the two cell lines respectively using QuickPrep micro mRNA purification kit (Pharmacia, United States) following the recommendations of the manufacturer.
SSH was performed by using PCR-Select TMcDNA subtraction kit (Clontech Laboratories Inc, United States) according to the recommendations of the manufacturer. The highly metastatic cell line SW620 was used as the tester while the lowly metastatic cell line SW480 was used as the driver in the forward hybridization, and vice versa in the reverse hybridization.
A total of 2 μg (4 μL) mRNA with 1 μL oligo (dT30) primer was heated to 70 °C for 2 min and rapidly cooled on ice. The reaction mixture was made up to 10 μL by adding 1 μL cDNA synthesis primer (10 μmol/L), 2 μL 5 × the first strand reaction buffer, 1 μL dNTP mix (10 mmol/L each) and 1 μL sterile H2O. Reverse transcription was started by adding 1 μL AMV reverse transcriptase (20 U/μL). The reaction mixture was incubated at 42 °C for 1.5 h to synthesize the first strand cDNA. The second strand cDNA synthesis was performed by immediately adding 48.4 μL sterile H2O, 16 μL 5 × the second strand buffer, 1.6 μL dNTP mix, 4 μL 20 × the second strand enzyme cocktail. The reaction mixture was incubated at 16 °C for 2 h. Double strand cDNA was blunted by adding of 2 μL T4 DNA polymerase and incubated at 16 °C for 20 min. The reaction was stopped by adding 20 × EDTA/glycogen mix into the reactive mixture. cDNAs were then extracted, precipitated, and resuspended in 50 μL of deionized water.
For the ligation of cDNA fragments, 43.5 μL of ds cDNA from the tester and driver was digested respectively with 1.5 μL of RsaI (10 U/μL) at 37 °C for 1.5 h. The reaction was terminated by adding 20 × EDTA/glycogen mix. The resulting fragments of cDNAs were extracted, precipitated and finally resuspended in 5.5 μL of deionized water. Adaptor1 and adaptor2R were ligated separately to 2 μL of RsaI digested tester cDNA with 1:6 dilution in the presence of T4 DNA ligase at 16 °C overnight followed by heating at 70 °C for 5 min to inactivate the ligase. In order to determine whether the ligation efficiency was high or not, 1 μL of adaptor1-ligated and adaptor2R-ligated cDNAs of the tester was diluted into 200 μL H2O respectively and amplified in two separate 50 μL reactions. One reaction used G3PDH3’, 5’ primer, while the other used G3PDH3’ primer and PCR primer1. PCR parameters were as follows: 30 cycles at 94 °C for 30 s, at 65 °C for 30 s and at 68 °C for 2.5 min. The products were examined by electrophoresis on a 20 g/L agarose/EB gel.
In the first hybridization, 1.5 μL of RsaI digested driver cDNA was mixed with 1.5 μL of diluted adaptor1- or adaptor2R-ligated tester cDNA. The samples were denatured at 98 °C for 1.5 min and immediately incubated in a thermal cycler at 68 °C for 8 h. In the second hybridization, two kinds of sample resulting from the first hybridization were mixed in the presence of a freshly denatured driver cDNA. The samples were incubated at 68 °C for 18 h. After 200 μL of dilution buffer was added, the samples were incubated for an additional 7 min. Analysis of the subtraction efficiency was carried out using PCR amplification of G3PDH in diluted subtracted cDNA versus unsubtracted cDNA. PCR was performed for 33 cycles at 94 °C for 30 s, at 60 °C for 30 s and at 68 °C for 2 min. The products were monitored on a 20 g/L agarose/EB gel for an aliquot which was removed from each reaction after 18, 23, 28, 33 cycles.
A 1 μL of diluted subtraction mixture was amplified with PCR primer1 in the primary PCR. The reaction mixture was incubated at 75 °C for 5 min to extend the adaptors and followed in turn at 94 °C for 25 s, 30 cycles at 94 °C for 10 s, at 64 °C for 30 s and at 71 °C for 1.5 min. The primary PCR mixture was diluted 10-fold and 1 μL from that was used in secondary PCR with nested PCR primer1 and primer2R. The conditions of the reaction were 15 cycles at 94 °C for 10 s, at 68 °C for 30 s and at 72 °C for 1.5 min. Eight μL products from each PCR reaction of the secondary PCR was analyzed on a 20 g/L agarose/EB gel.
The second PCR products were purified and their concentrations were measured by a spectrophotometer. The T/A cloning method was performed by using pGEM-T vector system I (Promega, United States) according to the recommendations of the manufacturer. The PCR product was cloned into the vector with a molar ratio of 6:1 (insert: vector). A 2 μL purified PCR products (25 ng/μL) was used in a 10 μL-ligation-reaction system including 5 μL 2× Rapid ligation buffer, 1 μL pGEM-T Easy Vector (50 ng/μL), 1 μL T4 DNA ligase (3Weiss units/μL) and 1 μLdeionized water. The mixture was incubated at 4 °C overnight. The positive control was made by using the control insert DNA from the kit instead of the PCR products. The background control was made without inserting any fragments. The recombinant plasmids were transformed respectively into the competent JM109 E-coli cell with the CaCl2 method. A 100 μL of transformants was grown on 8 cm × 8 cm agar plates containing 100 μg/mL ampicillin, 100 μmoL/L IPTG and 100 μg/mL X-gal at 37 °C for 20 h when the blue/white staining could be clearly distinguished. White clones were counted, inoculated into 3 mL of LB liquid medium containing ampicillin and shaken overnight at 37 °C. The resulting bacterial culture was directly used as PCR templates to amplify the inserts in 50 μL reaction mixture containing 10×PCR buffer 5.0 μL, Mg2+ (25 mmol/L) 3 μL, nested primer 1 (20 μmol/L) 1 μL, nested primer 2R (20 μmol/L) 1 μL, dNTP mix (10 mmol/L) 1 μL, Taq enzyme mix (5 U/μL) 0.5 μL and bacterial culture 0.5 μL. PCR consisted of an initial denaturation step at 94 °C for 5 min, followed by 25 cycles at 94 °C for 30 s, at 68 °C for 30 s and at 72 °C for 2 min each. The PCR products were analyzed on an 10 g/Lagarose/EB gel.
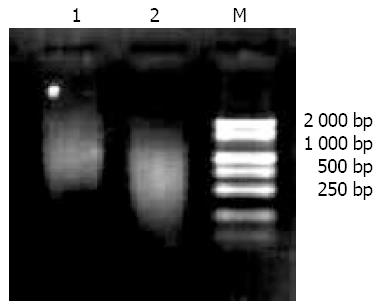
The extracted mRNA that was electrophoresed on 10 g/L agarose gel exhibited as a clear smear with over 0.5 kb length (Figure 1). Their absorbances(A260/A280 > 1.8) in spectrophotometry suggested that extracted mRNAs were in a highly purified quality.
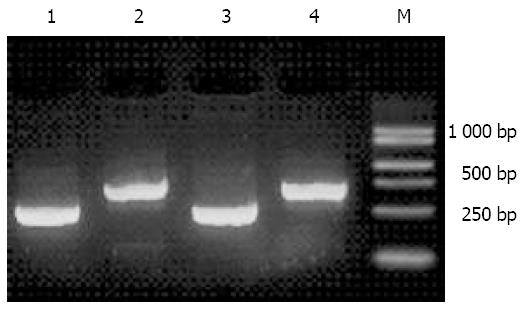
The results of 10 g/L agarose electrophoresis showed that cDNA before digested with RsaI, displayed a zonal smear with a length from 0.5 kb to 10 kb. After the digestion, the cDNA length became shorter with fragments from 0.1 kb to 2 kb, suggesting that cDNAs were completely digested (Figure 2).
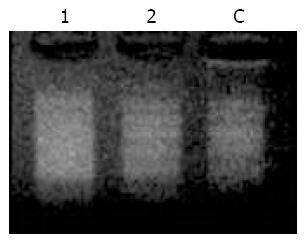
The high ligation efficiency was the most important factor for success of SSH. Figure 3 shows that the intensity of band 2/band1, band4/band3 was over 80%, indicating that subtracted cDNA library had the high ligation efficiency, 80% of the tester’ ds cDNA fragments were ligated with adaptors1 or 2R.
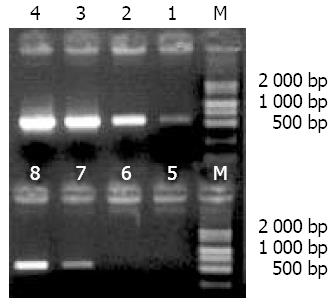
Nested primers from the inner sequences of adaptors were used to amplify the primary PCR products, and the smear was increased obviously after 15 cycles, in which some obscure bands with the lengths between 0.2 and 1.5 kb were observed (Figure 4).
Figure 5 shows that obvious bands were seen after 23 cycles in unsubtracted cDNA and after 33 cycles in diluted subtracted cDNA. The amount of G3PDH was significantly decreased after subtraction, indicating that the subtracted cDNA library had the high subtraction efficiency.
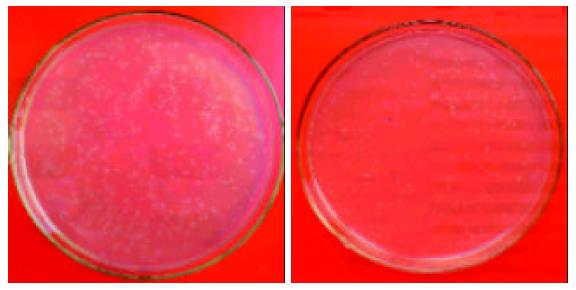
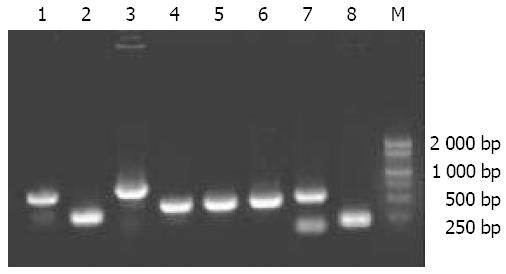
On the agar plates with 100 μL of transformants, 235 white clones were obtained in the forward hybridization (group A) while 232 white clones were obtained in the reverse hybridization (group R) (Figure 6). White clones accounted for more than 80% of the total clones. Ninety-eight percent and 91% of the white clones were demonstrated by PCR having the inserts with a length of 200-700 bp in groups A and R, respectively (Figure 7), indicating that a subtracted cDNA library specific for metastasis in CRC was successfully constructed.
Metastasis is the major cause of mortality in CRC patients. The molecular basis for metastasis has not been fully elucidated yet. Available studies have shown that many specific genes are involved in the process of metastasis, which may accelerate or suppress metastasis by a mechanism of cooperative or inhibitive interactions between genes[1]. Till now hundreds of genes have been reported to be involved in the regulation of metastasis in colorectal carcinoma. However, the existent methods for investigating the molecular mechanism of metastasis are mostly limited to the analysis of expressions of only one or several genes, which makes it difficult to understand the genes’ interactions and their relations to the metastasis.
One of the most important ways to understand the carcinogenesis and development of tumors is to clone and identify the differentially expressed genes. In recent years several cloning techniques for the differentially expressed genes have been established, including differential display PCR (DD-PCR), representational difference analysis (RDA), SSH and cDNA microarray. Until now, these techniques have been widely applied in the study of pathology[2], immune[3], embryo development[4], as well as the specific expression of tissue protein[5]. Although it is rapid, easy to perform and sensitive, DD-PCR has also some limitations, such as the higher false positive rate and the truncated length of obtained specific cDNA that is usually located in non-translated regions of 3’ poly (A). Likewise, RDA needs less amount of mRNAs samples and has lower false positive rate. But it is troublesome to perform more complicated steps of hybridization or PCR. Microarray-based methods have the potentials to revolutionize the study of differential gene expression under parallel conditions with higher automation. While a number of drawbacks of this approach have been found such as the higher expense and less sensitive than that of PCR-based techniques.
SSH was first put forward by Diatchenko et al in 1996. The basic principle was that common cDNAs in the paired materials were subtracted by subtractive hybridization and then suppressive PCR was carried out to amplify cDNA fragments specially expressed in tester. The advantage of SSH was the design of two different adaptors and the introduction of suppression PCR with the result that the differentially expressed cDNA fragments were amplified. It allows two subtractive hybridizations in the forward or reverse direction. Some mRNA expressed with low abundance could be detected, which provided clues for the further gene sequencing and identification[6-9]. With the development and improvement of SSH technique, it has become one of the most effective techniques for cloning differentially expressed genes. Since it was established, more than two hundred papers have been published concerning about carcinogenesis[10], tumor metastasis[11], signal transduction[12], apoptosis[13] and tumor therapy[14]. Combined with other molecular biological techniques such as DNA chip, it could clone tumor associated genes[15] and metastasis-associated genes in lung carcinoma, breast carcinoma and other malignancies[16-20]. Zhang et al[21] carried out SSH in human NCI-H69 cell line and its variant, a more aggressive NCI-N417 SCLC cell line, to characterize the variable expression of genes between the two cell lines, in which 42 genes were screened by the forward and reverse hybridizations. Wang et al[22] have identified five cDNA fragments with SSH that were expressed at much higher levels in the poorly metastatic cancer cell clone than in the highly metastatic variety. Until now studies on separating metastasis-associated genes in human CRC have not been reported.
In our study, SSH and bacterial culture PCR were performed to construct a subtracted cDNA library specific for the metastasis-related genes of human CRC, in which a pair of human colorectal carcinoma cell lines SW620 and SW480 was used. The SW480 cell line was derived from a Dukes’ stage B colon carcinoma that arose in a 50-year-old male patient with liver and mesenteric lymph node metastases. The SW620 cell line was derived from one of the lymph node metastases. The monoclonal origin of the two cell lines that was confirmed by the presence of shared maker chromosomes on cytogenetic analysis makes it suitable to study the genetic basis of their phenotypic differences. Hewitt et al.[23] have also found that the phenotypic differences of both SW620 and SW480 cell lines were retained, despite long-term culture in vitro.
Both the forward and reverse hybridizations were performed in this study. In the forward hybridization (group A), the highly metastatic cell line SW620 was treated as the tester, while the lowly metastatic cell line SW480 was treated as the tester in the reverse hybridization (group R). As a result, the total cDNAs in the tester and driver were subtracted efficiently. cDNA fragments appearing only in SW620 cells were amplified specially in group A and those only in SW480 cells were amplified in group R. After that, the second PCR products were inserted into T vectors and then screened by the blue/white screening system. Two hundred and thirty-five or 232 white clones were obtained in group A or group R respectively. The resultant subtractive cDNA library that was rich in metastasis-associated gene fragments of CRC was superior to that obtained by other methods for 4 reasons: (1) High specificity. Two subtractive hybridizations and two suppressive PCR were carried out to amplify the differentially expressed cDNA fragments, and suppress the amplification of unspecific cDNA fragments. Thus, the specificity of the subtractive library was greatly increased; (2) High sensitivity. The normalization of tester ss cDNA by SSH could make the differentially expressed genes with high or low abundance separated efficiently; (3) High efficiency. One SSH reaction could separate hundreds and thousands differentially expressed genes simultaneously; and (4) More information. The differentially expressed fragments in the forward hybridization might have a role in accelerating metastasis, and those in the reverse hybridization might be metastasis suppressor genes. The forward and reverse hybridizations could provide more information of the screening results.
However, the subtracted cDNA library constructed by SSH had some limitations. One of them was the requirement of large quantities of mRNA samples. The other was that differentially expressed cDNAs obtained from SSH were cDNA fragments digested by restrictive enzyme and needed to be amplified to get the full length. These drawbacks could be overcame by other methods such as the multi-cycle linear amplification based on cDNA synthesis and templet-guided in vitro transcription, in which enough mRNA for the continual research could be amplified with less than 1-50 ng RNA samples[24]. Thus, it is especially suitable for tissues that are hard to get or too little to provide enough RNAs. In addition, the full length of differential fragments could be obtained by the rapid amplification of cDNA ends (RACE)[25].
After the blue/ white screening, bacterial culture PCR was used in the present study to screen the positive recombinants. Compared with the routine methods, bacterial culture PCR was not only simple but also helpful for the further identifications. Our results showed that there were 98% clones with 200-700 bp inserts in group A and 91% positive clones of recombinants in group R, suggesting that these clones may contain metastasis-related cDNA fragments. The construction of this library is therefore of significance in screening and identifying the genes associated with metastasis of CRC, which would be beneficial to the elucidation of the molecular mechanism of metastasis and provide new gene targets for metastasis forecast, gene therapy and prognosis estimation in clinical practice.
Edited by Wang XL, Zhu L Proofread by Xu FM
| 1. | Welch DR, Rinker-Schaeffer CW. What defines a useful marker of metastasis in human cancer? J Natl Cancer Inst. 1999;91:1351-1353. |
| 2. | Sharma R, Samantaray S, Shukla NK, Ralhan R. Transcriptional gene expression profile of human esophageal squamous cell carcinoma. Genomics. 2003;81:481-488. |
| 3. | Dunphy JL, Balic A, Barcham GJ, Horvath AJ, Nash AD, Meeusen EN. Isolation and characterization of a novel inducible mammalian galectin. J Biol Chem. 2000;275:32106-32113. |
| 4. | Li P, Li JL, Hou R, Han QD, Zhang YY. [Changes in the gene expression profile of the left heart ventricle during growth in the rat]. Sheng Li Xuebao. 2003;55:191-196. |
| 5. | Ito H, Akiyama H, Iguchi H, Iyama K, Miyamoto M, Ohsawa K, Nakamura T. Molecular cloning and biological activity of a novel lysyl oxidase-related gene expressed in cartilage. J Biol Chem. 2001;276:24023-24029. |
| 6. | Sandhu H, Dehnen W, Roller M, Abel J, Unfried K. mRNA expression patterns in different stages of asbestos-induced carcinogenesis in rats. Carcinogenesis. 2000;21:1023-1029. |
| 7. | Zhu NX, Zheng S, Xu RZ, Gao RL, Shen JP, Yu RX. [Identification of multidrug resistance related genes in leukemia by suppression subtractive hybridization]. Zhonghua Xue Ye Xue Zazhi. 2003;24:14-17. |
| 8. | Ai JK, Huang X, Wang YJ, Bai Y, Lu YQ, Ye XJ, Xin DQ, Na YQ, Zhang ZW, Guo YL. [Screening of novel genes differentially expressed in human renal cell carcinoma by suppression subtractive hybridization]. Ai Zheng. 2002;21:1065-1069. |
| 9. | Shen C, Hui-Zhao D, Jiang G, Wang J, Zhang G. Molecular cloning, identification and analysis of lung squamous cell carcinoma-related genes. Lung Cancer. 2002;38:235-241. |
| 10. | Hufton SE, Moerkerk PT, Brandwijk R, de Bruïne AP, Arends JW, Hoogenboom HR. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 1999;463:77-82. |
| 11. | Asai T, Tomita Y, Nakatsuka S, Hoshida Y, Myoui A, Yoshikawa H, Aozasa K. VCP (p97) regulates NFkappaB signaling pathway, which is important for metastasis of osteosarcoma cell line. Jpn J Cancer Res. 2002;93:296-304. |
| 12. | Kostic C, Shaw PH. Isolation and characterization of sixteen novel p53 response genes. Oncogene. 2000;19:3978-3987. |
| 13. | Sakamoto H, Mashima T, Kizaki A, Dan S, Hashimoto Y, Naito M, Tsuruo T. Glyoxalase I is involved in resistance of human leukemia cells to antitumor agent-induced apoptosis. Blood. 2000;95:3214-3218. |
| 14. | Zhang Z, DuBois RN. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20:4450-4456. |
| 15. | Schlingemann J, Hess J, Wrobel G, Breitenbach U, Gebhardt C, Steinlein P, Kramer H, Fürstenberger G, Hahn M, Angel P. Profile of gene expression induced by the tumour promotor TPA in murine epithelial cells. Int J Cancer. 2003;104:699-708. |
| 16. | Yin J, Fan Q, Hao X, Fan D. [Identification of genes associated with human osteosarcoma metastasis suppression using suppression subtractive hybridization]. Zhonghua Yi Xue Yi Chuan Xue Zazhi. 2002;19:213-217. |
| 17. | Yin JN, Fan QY, Hao XB, Yang N. [Apoptosis antagonizing transcription factor is a gene highly expressed in highly metastatic human osteosarcoma cell lines]. Ai Zheng. 2002;21:127-131. |
| 18. | Koo TH, Lee JJ, Kim EM, Kim KW, Kim HD, Lee JH. Syntenin is overexpressed and promotes cell migration in metastatic human breast and gastric cancer cell lines. Oncogene. 2002;21:4080-4088. |
| 19. | Goldberg SF, Miele ME, Hatta N, Takata M, Paquette-Straub C, Freedman LP, Welch DR. Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63:432-440. |
| 20. | Devoogdt N, Hassanzadeh Ghassabeh G, Zhang J, Brys L, De Baetselier P, Revets H. Secretory leukocyte protease inhibitor promotes the tumorigenic and metastatic potential of cancer cells. Proc Natl Acad Sci USA. 2003;100:5778-5782. |
| 21. | Zhang L, Cilley RE, Chinoy MR. Suppression subtractive hybridization to identify gene expressions in variant and classic small cell lung cancer cell lines. J Surg Res. 2000;93:108-119. |
| 22. | Wang T, Lu Y, Liu S. [Suppression subtractive hybridization in the cloning of gene fragments in relation to lung cancer metastasis]. Zhonghua Zhong Liu Zazhi. 2001;23:296-300. |
| 23. | Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M, Stamp GW, Stetler-Stevenson WG. Validation of a model of colon cancer progression. J Pathol. 2000;192:446-454. |
| 24. | Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457-459. |
| 25. | Xu CS, Li YC, Lin JT, Zhang HY, Zhang YH. Cloning and analysing the up-regulated expression of transthyretin-related gene (LR1) in rat liver regeneration following short interval successive partial hepatectomy. World J Gastroenterol. 2003;9:148-151. |