Published online Mar 1, 2004. doi: 10.3748/wjg.v10.i5.638
Revised: December 9, 2003
Accepted: December 16, 2003
Published online: March 1, 2004
AIM: Most cancer cells acquire immortal capability by telomerase activation. The human telomerase reverse transcriptase gene (hTERT) is considered to be the major determinant of the enzymatic activity of human telomerase, and the hTERT promoter contains several c-Myc binding sites that mediate hTERT transcriptional activation. Few studies have examined the role of hTERT in hepatocarcinogenesis, and the relationship between c-Myc and telomerase in human hepatocellular carcinoma tissue is unknown.
METHODS: We measured hTERT mRNA levels and c-Myc oncoprotein expression in 57 patients with hepatocellular carcinoma using in situ hybridization and immunohistochemistry, respectively. The transcription regulation of hTERT was evaluated by transient transfection of pGL3-1375 into the human hepatocellular carcinoma cell line J5. To determine the relationship between c-Myc and the hTERT promoter, a 1375-bp DNA fragment encompassing the promoter was placed upstream of the luciferase reporter gene and transiently transfected into the cell line. Two additional hTERT promoter constructs (-776 and -100 bp region) and an hTERT promoter-LUC construct containing 2 c-Myc mutations (pGL3-181 MycMT) were also used for luciferase assays.
RESULTS: In 30 of 57 cases (52%), hTERT mRNA expression was associated with c-Myc protein expression. However, 16 of 57 cases (28%) showed strong hTERT mRNA detection without c-Myc protein expression, and 11 cases (19%) showed weak hTERT mRNA expression and strong c-Myc expression. Although luciferase activity was decreased between upstream 1375 bp and 776 bp, there was no significant difference between upstream 776 bp and 100 bp. Finally, there was no significant decrease in activity after transfection of the hTERT promoter-LUC construct.
CONCLUSION: The results indicate that c-Myc does not play a major role in gene regulation of the catalytic subunit of telomerase (hTERT) in human hepatocellular carcinoma. Other regulatory elements or epigenetic phenomena should be further investigated to understand hTERT gene regulation in human hepatocellular carcinoma.
- Citation: Chen CJ, Kyo S, Liu YC, Cheng YL, Hsieh CB, Chan DC, Yu JC, Harn HJ. Modulation of human telomerase reverse transcriptase in hepatocellular carcinoma. World J Gastroenterol 2004; 10(5): 638-642
- URL: https://www.wjgnet.com/1007-9327/full/v10/i5/638.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i5.638
Telomerase is a ribonucleoprotein enzyme that synthesizes G-rich telomeric repeats using its complementary RNA sequence as a template[1,2]. Telomerase is expressed in most human cancers and immortal cell lines but is inactive in normal somatic cell lines or tissue[3-5]. Recent reports support the concept that activation of telomerase may be an important and obligate step in the development of most malignant tumors[6,7], including human hepatocellular carcinoma (HCC)[8]. The human telomerase catalytic subunit (hTERT) has been shown to be a rate-limiting determinant of the enzymatic activity of human telomerase[9,10]. Takakura et al[11] identified the proximal 181-bp core promoter region essential for transactivation of hTERT. Their findings suggest that hTERT expression is strictly regulated at the transcription machinery, and that the proximal core promoter containing an E-box which binds to Myc/Max, as well as the 3’-region containing the GC-box which binds to Sp1, is required for transactivation of hTERT[12]. Their findings further indicate that c-Myc and Sp1 cooperatively function as the major determinants of hTERT expression, and that the switching functions of Myc/Max and Mad/Max might also play roles in telomerase regulation. Wang et al[13] added further support that Myc induce telomerase both in normal human mammary epithelial cells and in normal human diploid fibroblasts by introducing HPV-16, E6 protein into these cells. Their findings suggest that the ability of c-Myc to activate telomerase may contribute to its ability to promote tumor formation. Further, telomerase activity in estrogen receptor-positive MCF-7 cells was upregulated by treatment with 17 β-estradiol. Kyo et al[14] reported that estrogen activated c-Myc expression in MCF-7 cells, and that E-boxes in the hTERT promoter that bind to c-Myc/max played additional roles in estrogen-induced transactivation of hTERT.
By using TRAP assay, we previously measured telomerase activity in surgically resected specimens from 25 cases of hepatocellular and adjacent healthy tissues[15]. Telomerase activity was detected in 21 of the 25 HCC specimens from 25 different cases. This telomerase activity was correlated with human telomerase reverse transcriptase (hTERT) mRNA isoform expression but was poorly related to c-Myc expression in the hepatoma cell line J5[16]. However, the role of c-Myc in hTERT expression in HCC remains unresolved.
In this study, we explored the relationship between hTERT mRNA regulation and c-Myc expression by RNA in situ hybridization and immunohistochemistry stain, respectively. The methods of in situ hybridization and immunohistochemistry are semiquantitative and can determine localization. In addition, to determine the cis-elements essential for transcriptional activation of hTERT, luciferase assays were performed with reporter plasmids with serial deletions or mutation of the core promoter using hepatoma cell line J5. The results provide evidence for a role of c-Myc in the regulation of hTERT in hepatoma cells.
All culture media including fetal bovine serum were purchased from Gibco Laboratories (Grand Island, NY). L-glutamine and penicillin/streptomycin were obtained from Sigma (St. Louis, MO).
WI38 cells (normal human fibroblasts) were obtained from the American Type Culture Collection and grown in DMEM containing 2 mmol/L L-glutamine, 50 U/mL penicillin, 50 mg streptomycin, and 100 mL/L fetal bovine serum. J5[16] was maintained in RPMI 1640 medium containing 3 g/L L-glutamine and penicillin/streptomycin. All cell lines were cultivated in an atmosphere of 50 mL/L CO2 at 37 °C.
Total RNA was obtained from the HT29 cells (ATCC, Rockville, MD) by addition of TRIZOL reagent (Life Technologies, Rockville, MD) according to the manufacturer’s instructions.
Ten micrograms of total RNA were used as a template for cDNA synthesis with Moloney murine leukemia virus (M-MTV) reverse transcriptase and oligo (dT)12-18 (SUPERSCRIPT Preamplification System, Life Technologies). Subsequently, the forward primer 5’-CGG AAG AGT GTC TGG AGC AA-3’ and the reverse primer 5’-GGA TGA AGC GGA GTC TGG-3’ were designed for amplification of a 145-bp segment of hTERT spanning from nucleotide position 1784 to 1928 (GenBank accessory No. AF0 15950). Thirty-five PCR cycles were performed. For each cycle, the sample was denatured at 94 °C for 30 s, annealed at 55 °C for 60 s, and extended at 72 °C for 60 s. A 10 µL sample from 100 µL PCR solution was fractionated by electrophoresis on 20 g/L agarose gel. Subsequently, the PCR product was eluted from the agarose gel and subcloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA), generating the construct-designated pCRII/hTERT-145.
The sense and antisense riboprobes were synthesized from Bam HI- and Eco RV-linearized PCRII/hTERT-145 according to the manufacturer’s instructions using T7 and SP6 RNA polymerase, respectively, and labeled with digoxigenin-UTP (DIG RNA Labeling Kit, SP6/T7, Roche Molecular Biochemicals, Mannheim, Germany). Moreover, the housekeeping gene GAPDH was used to confirm the presence of intact RNA within the slides from each sample used for ISH.
Formalin-fixed, paraffin-embedded tissue sections (4-µm thick) were deparaffinized with two 10 min washes with xylene and a graded series of alcohols for 3 min each. The deparaffinized tissues were then pretreated with 20 µg/mL proteinase K (Sigma) and 40 µg/mL pronase (Roche Molecular Biochemicals) at room temperature for 30 min. The tissues were then fixed with 40 g/L paraformaldehyde (Sigma) in phosphate-buffered saline at room temperature for 10 min and then acetylated with 2.5 mL/L acetic anhydride in 0.1 mmol/L triethanolamine-HCl (pH8.0) at room temperature for 10 min.
Prehybridization was carried out in hybridization solution containing 500 g/L deionized formamide (Merck, Darmstadt, Germany), 5 × SSC (1 × SSC = 150 mmol/L NaCl, 15 mmol/L sodium citrate, pH7.2), 1 g/L N-lauroylsarcosine 2 g/L sodium dodecylsulfate, 20 mL/L blocking solution (DIG wash and block buffer set, Roche Molecular Biochemicals) and 250 µg/mL sonicated salmon sperm DNA (Invitrogen) at 50 °C for 1 h. The slides were then incubated in a moist chamber at 50 °C for 16 h with the hybridization solution containing 0.1 to 0.5 µg/mL digoxigenin-labeled RNA probe. The slides were subsequently washed twice with 50% formamide-2 × SSC at 50 °C for 30 min, twice with 2 × SSC at room temperature for 15 min, and twice with 0.2 × SSC at room temperature for 15 min. The slides were then equilibrated with 1× washing solution for 2 min and incubated with 10 mL/L blocking solution (DIG wash and block buffer set, Roche Molecular Biochemicals) for 10 min.
The tissues were incubated with a sheep monoclonal antidigoxigenin antibody (Roche Molecular Biochemicals) diluted 1100 in 10 mL/L blocking solution at room temperature for 2 h. After washed three times with 1 × washing solution (Roche Molecular Biochemicals), the color reaction was carried out by incubation with 1 × nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate solution (Roche Molecular Biochemicals) at room temperature overnight. The slides were then counterstained with nuclear fast red for 5 min and then mounted with Crystal Mounting reagent (DAKO, Glostrup, Denmark).
Two independent observers evaluated the signal intensity of hTERT expression, which was semiquantitated as strong, moderate, weak, or no staining. Sense and antisense probes were applied to paired serial slides, and the noncoding strand detected by sense probes was used as a negative control.
Immunohistochemical staining was performed to determine the expression of c-Myc. The immunostaining procedure was performed using the labeled streptavidin-biotin method (LASB-2 Kit, DAKO). Briefly, the tissue was placed in a boiling citrate buffer (pH6; ChemMateTM, DAKO) twice for 5 min in a microwave oven at 750 W after deparaffinization and rehydration, as previously described. Quenching of the endogenous peroxidase activity by incubation with 30mL/L hydrogen peroxide for 10 min at room temperature was followed by incubation with mouse monoclonal antibody NCL-cMYC (Clone 9E11, Novocastra Laboratories Ltd., Newcastle-upon-Tyne, UK) diluted 1:200 at room temperature for 2 h. After washed with Tris-buffered saline containing 1 g/L Tween-20, the specimens were sequentially incubated for 10 to 30 min with biotinylated anti-mouse immunoglobulins and peroxidase-labeled streptavidin. Staining was performed after 10 min of incubation with a freshly prepared substrate-chromogen solution containing 3% 3-amino-9-ethylcarbazole and hydrogen peroxide. Finally, the slides were lightly counterstained with hematoxylin, washed with water and then mounted. Two independent observers assessed the sections. Because the extent of c-Myc labeling index was heterogeneous, the scoring system included both the staining intensity and the percentage of stained cells[17]. Staining intensity was graded as no staining (0), weak (1), moderate (2), or strong (3). The percentage of tumor cells with c-Myc staining was scored as follows: 1, < 5%; 2, 5% - 20%; 3, 21% - 50%; 4, > 50%. The multiplication values were then grouped into 4 scores as 0 (multiplication values 0, 1), 1 (multiplication values 2, 3), 2 (multiplication values 4, 6), or 3 (multiplication values 8, 9, 12).
One microliter of genomic DNA was obtained as DNA template for use in PCR amplification of the hTERT promoter. The forward primer 5’-CCC ACG CGT GCA TTC GTG GTG CCC GGA GC-3’ and the reverse primer 5’-CCC AGA TCT ATC GCG GGG GTG GCC GGG GCC AGG-3’ were designed on the basis of a published hTERT promoter sequence[11]. The PCR product was amplified in the presence of 1 µmol primers with Taq DNA polymerase (Takara Shuzo Company, Shiga, Japan) for 35 cycles of 1 min at 95 °C, 1 min at 56 °C, and 1 min at 72 °C. DNA sequencing using the reverse primer was performed directly from the gel-purified PCR product or individual PCR product subcloned into the pCRII-TOPO vector. The analysis of the DNA sequences was compared with the wild-type sequence.
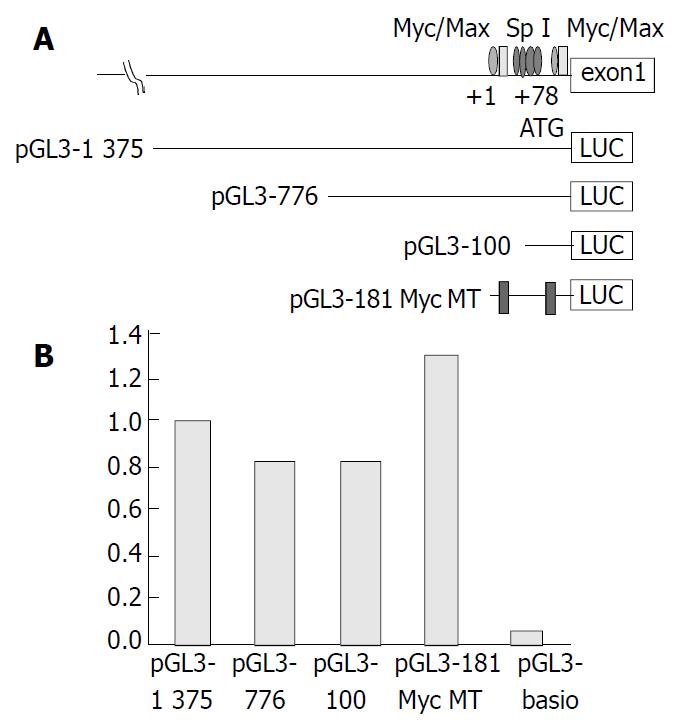
A P-3996 construct containing 3996 bp of sequences upstream of the ATG (with A being position 1) plus exon 1 (219 bp), intron 1 (104 bp), and 37 bp of exon 2 of hTERT was used (a kind gift from Silvia Bacchetti, Department of Pathology and Molecular Medicine, McMaster University, Canada)[18]. Various lengths of DNA fragments upstream of the initiating ATG codon were PCR amplified and inserted into luciferase reporter vector pGL3-Basic, a promoter- and enhancerless vector (Promega, Madison, WI) in sense orientation relative to the luciferase coding sequence at MluI and BglII sites. The sequences of primers were as follows: pGL3-1375-forward: 5’-CCCACGCGTAGACAATTCACAAACACAGC-3’, pGL3-776-forward: 5’-CCCACGCGTGCCAGCAGGA GCGCCTGGCT-3’, pGL3-100-forward: 5’-CCCACGCG TCCGCGCGGACCCCGCCCCGT-3’ and reverse (common): 5 ’ - CC C AG A TC TA T CG CG G GG GT G GC C GG GG C CAGGGCTTC-3’ with the PCR condition supported by Satoru Kyo (Department of Obstetrics and Gynecology, Kanazawa University, School of Medicine, Ishikawa, Japan)[11]. The PCR product was amplified in the presence of 1 µL primers with TaKaRa Taq DNA polymerase (Takara Shuzo Company, Shiga, Japan) for 30 cycles of 30 s at 96 °C, 45 s at 62 °C, and 7 min at 72 °C, and 30 min at 72 °C. The products were confirmed to have correct sequences by nucleotide sequencing, and their quantity and quality were routinely checked by agarose gel electrophoresis. All plasmid DNAs were purified with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany).
Transient transfection of luciferase reporter plasmids was performed using LipofectAMINE 2000 (LF2000, Invitrogen), according to the protocol recommended by the manufacturer. In brief, 5 × 104 cells were seeded on 24-well plates, cultured overnight, and exposed to transfection mixtures containing 2 mg luciferase reporter plasmids for 4 h at 37 °C. Then, 0.5 ml growth media was added and cells were harvested 48 h after transfection. Luciferase assays were performed with the dual-luciferase reporter assay system (Promega) according to the manufacturer’s protocols. The pGL3-control plasmid (1 mg/well, Promega) was also transfected into each cell line for better comparison among cell lines with different transfection efficiencies. The pRL-SV40 (1 ng/well, Promega) containing the Renilla reniformis luciferase gene was cotransfected with the hTERT promoter-luciferase constructs (1 mg/well) for normalization of the luciferase activity in each transfection. The MLX microtiter plate luminometer (Dynex Technologies, Chantilly, VA) was used to detect luciferase activity. All experiments were performed at least 3 times in each plasmid and represented the average relative luciferase activity.
To evaluate the relationships among paired groups, the Fisher exact test was performed using SPSS 10.0 software. Additionally, the correlation of paired groups was analyzed using chi-square test with the SPSS program. A P value ≤ 0.05 was considered statistically significant.
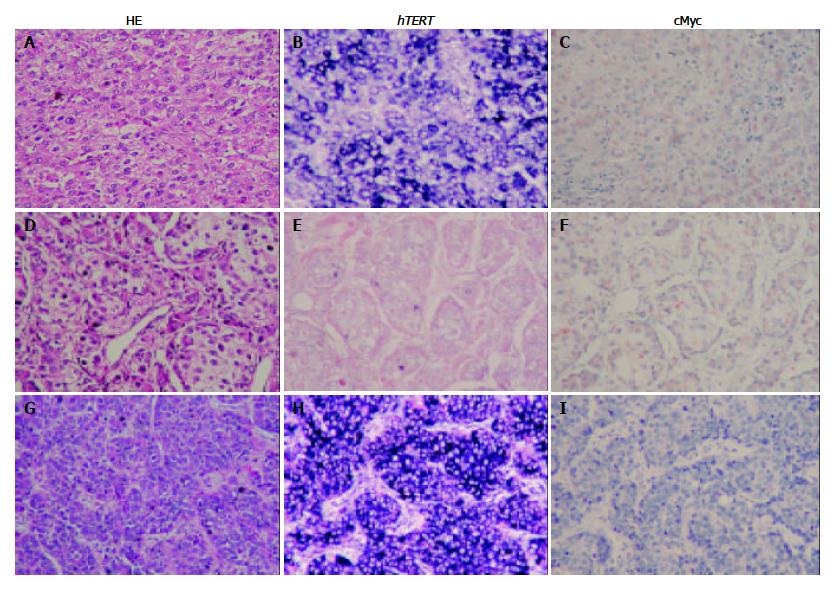
To investigate hTERT expression in HCC tissue, in situ hybridization was applied. Immunohistochemistry stain was used to observe c-Myc expression and its relationship with hTERT mRNA. Forty-seven of 57 cases showed weak to strong hTERT mRNA expression. The expression of hTERT mRNA was not related to tumor differentiation (P < 0.815) (Table 1). Forty-three of 57 cases showed c-Myc expression without tumor differentiation (P < 0.348) (Table 2).
| Differentiation | hTERT expression | Number | |||
| None | Weak | Moderate | Strong | ||
| Strong | 1 | 2 | 2 | 7 | 12 |
| Moderate | 7 | 3 | 4 | 14 | 28 |
| Weak | 2 | 4 | 3 | 8 | 17 |
| Numbe | 10 | 9 | 9 | 29 | 57 |
| Differentiation | c-Myc expression | Number | |||
| 0 | 1 | 2 | 3 | ||
| Strong | 3 | 0 | 2 | 7 | 12 |
| Moderate | 5 | 7 | 7 | 9 | 28 |
| Weak | 6 | 3 | 2 | 6 | 17 |
| Numbe | 14 | 10 | 11 | 22 | 57 |
Thirty of 57 cases (52%) of hTERT mRNA expression were associated with c-Myc protein expression (30/57). However, 16 of 57 cases (28%) showed strong hTERT mRNA detection with no Myc protein expression, whereas 11 of 57 cases (19%) showed weak hTERT mRNA expression with strong c-Myc detection (P < 0.079) (Figure 1).
Three different-length DNA fragments (-1375, -776, and -100 bp) encompassing the hTERT promoter were placed upstream of the luciferase reporter gene, as was an hTERT promoter-Luc construct containing 2 c-Myc mutations (pGL-181 MycMT, a gift from Kyo et al[12]). All constructs were transiently transfected into HCC cell line J5 for luciferase study. Luciferase activity decreased between upstream 1375 and 776 bp, but there was no significant difference of luciferase activity between upstream 776 and 100 bp or the 2 c-Myc mutations (Figure 2).
In a previous report, we demonstrated that telomerase activity in the HCC cell line J5 was not related to c-Myc expression[16]. To our knowledge, this is the first study to determine the role of c-Myc in hTERT HCC. According to in situ hybridization and immunohistochemistry analysis, only half of hTERT mRNA expression co-occurred with c-Myc protein expression. Twenty-eight percent of HCC tissue samples had strong hTERT mRNA detection with no or weak c-Myc protein expression, 19% of HCC tissue samples had no or weak hTERT mRNA expression with strong c-Myc expression. However, several studies reported that Myc expression could transactivate hTERT via 2 E-boxes in cooperation with Sp1 motif. One of our constructs (a gift from Kyo), which encompassed 4 Sp1 and 2 c-Myc mutations, showed a high luciferase activity in the HCC cell line.
In contrast to the data of Kyo et al[14], pGL3-181MycMT, a double c-Myc mutant, compared with wild type pGL3-181, exhibited a 50% decreased luciferase activity when transfected to the MCF-7 breast cell line. These results implicate that c-Myc is a positive regulator of hTERT, though other yet undetermined regulatory elements of hTERT in HCC may exist. For example, hepatitis B virus pre-S2/S gene has been found to be a cis-activator of the hTERT promoter[19]. By transfection of HBx gene into the HepG2 cell line, the activity of telomerase and apoptosis were decreased[20]. Further investigation of non-c-Myc regulatory proteins in hepatoma is required in the future.
In our 16 HCC tissue specimens and 1 J5 cell line, hTERT promoter cis-element sequencing was performed. There was a polymorphism site (A transversion to T) just 3 bp away from the distal E-box, which might have affected the binding affinity of c-Myc[21]. This effect might explain why two E-box mutations still had a high telomerase activity. However, more evidence is required in support of this polymorphism nucleotide in 2 E-box mutation construct.
Furthermore, the presence of a large CpG island with a dense CG-rich content implicates that DNA methylation and chromatin structure may play a role in the regulation of hTERT expression. Devereux et al[22] demonstrated that the promoter of one hTERT-negative fibroblast cell line, SUSM-1, was methylated at all sites examined. Treatment of SUM-1 cells with the demethylating agent induced the cells to express hTERT, suggesting a potential role for DNA methylation in negative regulation. This epigenetic mechanism could explain why 19% of HCC samples showed strong c-Myc detection with no or weak hTERT mRNA expression. The role of GC island methylation in the regulation of hTERT expression merits further study.
In the Kyo et al[12] report, the cis-acting effect of E-boxes and the Myc or Max requirement for transactivation varied among different cell types. Deletion and mutation of the E-box resulted in significant loss of transcriptional activity in C33A cells, but not in SiHa cells. In C33A cells, expression of Myc and Max had only marginal effects on transactivation. This diversity among different cell lineages suggests a varied role of c-Myc in hTERT gene regulation. The data also indicate that there are multiple levels of regulation of hTERT activity in human neoplasm.
In summary, in the present hepatoma tissue study, 50% of hepatomas showed c-Myc overexpression with hTERT transcript upregulation. Other regulator elements and epigenetic mechanisms may be involved in hTERT transcript regulation. The proximal c-Myc motif plays a minor role in hTERT gene regulation. The results of immunohistochemistry and promoter-constructed luciferase analyses suggest that, in HCC, hTERT regulation is not restricted to c-Myc and involves other mechanisms.
Edited by Wang XL Proofread by Zhu LH
| 1. | Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622-6626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1496] [Cited by in RCA: 1603] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 2. | Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 279] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 3. | Counter CM, Hirte HW, Bacchetti S, Harley CB. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci USA. 1994;91:2900-2904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 465] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 4. | Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011-2015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5156] [Cited by in RCA: 5233] [Article Influence: 168.8] [Reference Citation Analysis (0)] |
| 5. | Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 2003] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 6. | Chadeneau C, Hay K, Hirte HW, Gallinger S, Bacchetti S. Telomerase activity associated with acquisition of malignancy in human colorectal cancer. Cancer Res. 1995;55:2533-2536. [PubMed] |
| 7. | Zhan WH, Ma JP, Peng JS, Gao JS, Cai SR, Wang JP, Zheng ZQ, Wang L. Telomerase activity in gastric cancer and its clinical implications. World J Gastroenterol. 1999;5:316-319. [PubMed] |
| 8. | Tahara H, Nakanishi T, Kitamoto M, Nakashio R, Shay JW, Tahara E, Kajiyama G, Ide T. Telomerase activity in human liver tissues: comparison between chronic liver disease and hepatocellular carcinomas. Cancer Res. 1995;55:2734-2736. [PubMed] |
| 9. | Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, Weinberg RA. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 308] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 10. | Kim HR, Christensen R, Park NH, Sapp P, Kang MK, Park NH. Elevated expression of hTERT is associated with dysplastic cell transformation during human oral carcinogenesis in situ. Clin Cancer Res. 2001;7:3079-3086. [PubMed] |
| 11. | Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551-557. [PubMed] |
| 12. | Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Ariga H, Inoue M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 2000;28:669-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 380] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 13. | Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769-1774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 475] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 14. | Kyo S, Takakura M, Kanaya T, Zhuo W, Fujimoto K, Nishio Y, Orimo A, Inoue M. Estrogen activates telomerase. Cancer Res. 1999;59:5917-5921. [PubMed] |
| 15. | Hsieh HF, Harn HJ, Chiu SC, Liu YC, Lui WY, Ho LI. Telomerase activity correlates with cell cycle regulators in human hepatocellular carcinoma. Liver. 2000;20:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Chen CJ, Tsai NM, Liu YC, Ho LI, Hsieh HF, Yen CY, Harn HJ. Telomerase activity in human hepatocellular carcinoma: parallel correlation with human telomerase reverse transcriptase (hTERT) mRNA isoform expression but not with cell cycle modulators or c-Myc expression. Eur J Surg Oncol. 2002;28:225-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Brabletz T, Herrmann K, Jung A, Faller G, Kirchner T. Expression of nuclear beta-catenin and c-myc is correlated with tumor size but not with proliferative activity of colorectal adenomas. Am J Pathol. 2000;156:865-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 118] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 365] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 19. | Horikawa I, Barrett JC. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J Natl Cancer Inst. 2001;93:1171-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Zhou W, Shen Q, Gu B, Ren H, Zhang D. [Effects of hepatitis B virus X gene on apoptosis and the activity of telomerase in HepG(2) cells]. Zhonghua Ganzangbing Zazhi. 2000;8:212-214. [PubMed] |
| 21. | O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087-6090. [PubMed] |