Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3194
Revised: April 23, 2004
Accepted: April 29, 2004
Published online: November 1, 2004
AIM: To study the difference of gene expression profile changes in Barrett’s esophagus (BE) and cardia intestinal metaplasia (CIM) and to screen the novel genes in the early stage by cDNA microarray.
METHODS: cDNA retrotranscribed from an equal amount of mRNA from BE and CIM epithelial tissues was labeled with Cy3 and Cy5 fluorescence as probes. The mixed probe was hybridized with three pieces of BiostarH-40 s double dot human whole gene chip. The chips were scanned with a ScanArray 4000. The acquired images were analyzed using GenePix Pro 3.0 software.
RESULTS: A total of 141 genes were screened out that exhibited different expression in all three chips. There were 74 upregulated and 67 downregulated genes in gene expression profiles of BE which were two times of that in CIM.
CONCLUSION: There is a difference in gene expression level between BE and CIM epithelia. These 141 genes probably relate to the occurrence and development of BE and the progression to adenocarcinoma.
- Citation: Chang Y, Gong J, Liu B, Zhang J, Dai F. Gene expression profiling in Barrett’s esophagus and cardia intestinal metaplasia: A comparative analysis using cDNA microarray. World J Gastroenterol 2004; 10(21): 3194-3196
- URL: https://www.wjgnet.com/1007-9327/full/v10/i21/3194.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i21.3194
The incidence of adenocarcinoma in the esophagus and gastroesophageal junction (GEJ) has been increasing over the last two decades in North America and Europe[1]. Barrett’s esophagus (BE) is thought to be a premalignant condition for esophageal adenocarcinoma and most of adenocarcinomas at GEJ[2]. Recently the presence of cardia intestinal metaplasia (CIM) in some normal GEJ has been described[3]. The relation of this condition to BE has not yet been investigated.
Recently, cDNA microarray methods are applied to the study of gene expression. The differentially expressed genes in different specimens may be detected with parallel analysis by gene chips which has greatly improved the traditional experiments in that only a single or several gene expression can be observed for each test, thereby speeding up the identification of differentially expressed genes and the construction of differential expression profiles.
This study was conducted on three 4096-chips, to analyse the gene expression profiles between BE and CIM epithelia, and to screen novel genes by cDNA microarray, which is helpful to understand the molecular mechanism of cell transformation and provides molecular markers and target genes for clinical diagnosis, prevention and treatment of BE and esophageal adenocarcinoma and adenocarcinoma at GEJ.
Tissue specimens in this study were provided by the Second Hospital of Xi’an Jiaotong University, with the approval of hospital and personnel authorities. BE and CIM tissues from 13 operated or biopsy specimens (7 BE and 6 CIM) were proved pathologically. For each sample, one part was cut and frozen in liquid nitrogen immediately after resection, and the other part was used for histopathological examination. Standardized endoscopy was performed by experienced endoscopists. The appearance of sternoclavicular joint (SCJ) was carefully studied from both prograde and retrograde views. BE was defined as any columnar-lined mucosa above the GEJ, which was further confirmed by Alcian blue staining. CIM was defined by the presence of barrel-shaped goblet cells in normal GEJ.
4 086 of target cDNA clones used in cDNA chips were provided by United Gene Ltd. and cooperative fellows. These genes were amplified with PCR using universal primers and then purified with standard method. The quality of PCR was monitored by agarose gel electrophoresis. The obtained genes were dissolved in 3 × SSC spotting solution and then spotted on silylated slides (Telechem. Inc) by Cartesian 7 500 Spotting Roboter (Cartesian, Inc). Each target gene was dotted twice. After spotting, the slides were hydrated for 2 h and dried for 0.5 h at room temperature. The samples were cross-linked with UV light and treated with 2 g/L SDS, H2O and 2 g/L NaNBH4 for 10 min respectively. Then the slides were dried in cold condition and ready for use.
Total sample RNA was extracted by single step method. Briefly, after taken out from liquid nitrogen, specimens were ground into tiny powder while liquid nitrogen was added in ceramic mortar and then homogenized in D solution plus 10 mL/L mercaptoethanol. After centrifugation, the supernatant was extracted with phenol: Chloroform (1:1), NaAc and acidic phenol: Chloroform (5:1) respectively. The aqueous phase was precipitated by an equal volume of isopropanol and centrifuged. The precipitates were dissolved with purified H2O. After further purification by LiCl precipitating method, the obtained RNA sample was analyzed. mRNAs were isolated and purified with Oligotex mRNA Midi Kit (Quagen, Inc.). The fluorescence-labeled cDNA probe was prepared through retrotranscription, referring to the method of Schena. The probes from CIM tissue were labeled with Cy3-dUTP, while those from BE tissue with Cy5-dUTP respectively. The probes were mixed and precipitated by ethanol, and then resolved in 20 mL hybridization solution (5 × SSC + 2 g/L SDS).
Probes and chips were denatured respectively in 95 °C bath for 5 min, then the probes were added on the chip. They were hybridized in a sealed chamber at 60 °C for 15-17 h and washed in turn with solutions of 2 × SSC + 2 g/L SDS, 0.1 × SSC + 2 g/L SDS and 0.1% SSC for 10 min each, then dried at room temperature.
The chips were read by Scan Array 4000 Scanner (General Scanning Inc). The overall intensities of Cy3 and Cy5 were normalized and corrected by a coefficient according to the ratios of the 40 located housekeeping genes. The acquired image was further analyzed by GenePix Pro 3.0 software with a digital computer to obtain the intensities of fluorescent signals and the Cy3/Cy5 ratio. The data were taken on an average of the two repeated spots. The differentially expressed genes were defined as follows: The absolute value of the Cy5/Cy3 natural logarithm was more than 0.69 (the variation of gene expression was more than 2-fold). Either Cy3 or Cy5 signal value was required for more than 800. The PCR results were satisfactory.
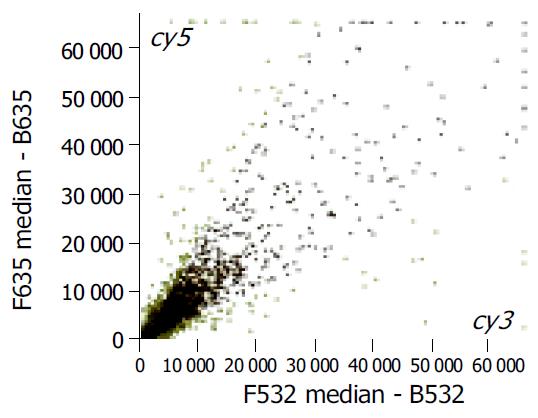
The scatter plots that were plotted with Cy3 and Cy5 fluorescent signal values displayed a quite disperses pattern in distribution. Most of the spots gathered around a 45° line, in which red spots represented the area where the signal intensities varied between 0.5 to 2-fold compared with those of the control. Some yellow spots distributed beyond or far from 45° line indicated the existence of abnormal gene expressions in BE and CIM epithelia. Their signal intensities were 2 times more than that of the control (Figure 1).
As shown in Figure 2, 141 genes were screened out that exhibited different expressions in all three chips, there were 74 up-regulated and 67 down-regulated genes in the gene expression profiles of BE which was 2 times of that in CIM. These genes might be divided into 16 groups (Table 1) according to their functions.
| Gene function | n | Ratio > 2.0 | Ratio < 0.5 |
| Proto-oncogene and tumor suppression genes | 9 | 6 | 3 |
| Cell signals and transducing proteins | 5 | 2 | 3 |
| Cell cycle proteins | 8 | 6 | 2 |
| Extra-pressure reaction proteins | 1 | 1 | 0 |
| Cell regulatory proteins | 4 | 4 | 0 |
| Cell apoptosis related proteins | 3 | 2 | 1 |
| DNA synthesis, repair and recombinant proteins | 3 | 2 | 1 |
| DNA binding, transcription and its factor | 4 | 3 | 1 |
| Cell receptors | 1 | 0 | 1 |
| Cell surface antigen and adhesion proteins | 10 | 4 | 6 |
| Ion-channel and transporters | 11 | 6 | 5 |
| Metabolism-related proteins | 16 | 7 | 9 |
| Protein synthesis-related genes | 11 | 6 | 5 |
| Development-related genes | 0 | 0 | 0 |
| Other genes | 38 | 17 | 21 |
| New genes | 17 | 8 | 9 |
| Total | 141 | 74 | 67 |
Over the last two decades, the incidence of adenocarcinoma of the esophagus and gastric cardia has been increasing rapidly. Barrett’s metaplasia is recognized as a precancerous lesion of esophageal adenocarcinoma and most of the adenocarcinomas were found at the GEJ. Progression from metaplasia, dysplasia to adenocarcinoma has been well understood[4]. Traditionally Barrett’s esophagus (BE) is defined as a circumferential segment of columnar lined epithelium of 2 or 3 cm in length in the lower esophagus. Recently this macroscopic definition has been questioned, as it excludes shorter segments and “tongues” of columnar lined epithelium, which are frequently found in the distal esophagus, and endoscopic measurements may be imprecise. It has therefore been proposed that the diagnosis of BE be reserved for patients with intestinal metaplasia detected in biopsy specimens from the distal esophagus. Recently the presence of cardia intestinal metaplasia (CIM) in some normal GEJ has been described[5-7]. Detection of intestinal metaplasia in the distal esophagus as well as within the gastric cardia was reported with increasing frequency[8]. The prevalence of BE was reported to vary from 2% to 12% and that of CIM from 5% to 23% in patients undergoing routine upper endoscopy[9]. The detection of intestinal metaplasia in the BE potentially committed patients to regular surveillance with biopsy. The incidence of adenocarcinoma in patients with BE was estimated to be 30-50 times that of the general population[10,11]. However, the exact incidence of cancer in patients with BE is unknown, and the role of CIM as a premalignant lesion is still unclear. The relation of this condition to BE has not yet been investigated. It is, however, not clear whether intestinal metaplasia of the cardia and esophageal mucosa origins has a common pathogenesis and identical risk factors. Despite the frequent occurrence of cardia intestinal metaplasia and its association with H pylori gastritis and multifocal gastric intestinal metaplasia, there is still no evidence that this finding could indicate an increased risk of malignancy in the cardia. The well known association of traditional BE with symptoms and endoscopic features of gastroesophageal reflux disease (GERD) has, however, not been confirmed in CIM[12], Future studies should differentiate BE from CIM in order to enhance our understanding of the pathophysiology and the malignant potential of each clinical entity. It is therefore necessary to explore new and efficacious diagnostic methods to discriminate BE from CIM.
cDNA microarray methods have been applied in the study of gene expression, DNA sequence, novel genes and gene mutants, DNA polymorphism, and in screening drugs, diagnosing diseases and mapping gene library[13]. The differentially expressed genes in different specimens may be detected with parallel analysis by gene chips which has greatly improved the traditional experiments in that only a single or several gene expression can be observed for each test, thereby speeding up the identification of differentially expressed genes and the construction of different expression profiles. Profiling of differentially expressed genes of BE to each of the normal upper gastrointestinal (GI) mucosae, including gastric, duodenal, and squamous epithelia of the esophagus by cDNA expression array was also reported[14]. It has been shown that there was a clear distinction among the expression profiles of gastric, duodenal, and squamous epithelia whereas the BE profiles showed a considerable overlap with normal tissues. Furthermore, the clusters of genes that are specific to each of the tissues from BE, and a cluster of genes distinct from squamous and non-squamous epithelia, were identified. However, no investigation on the difference in gene expression profiles between BE and CIM epithelia by gene chip has been reported yet.
In the present study, we performed an analysis on three 4096 chips in order to acquire the difference in gene expression profiles between BE and CIM epithelia. The results showed that a total of 141 genes were screened out that exhibited different expressions in all three chips. In the gene expression profiles of BE there were 74 upregulated and 67 downregulated genes which were two times of those of CIM. A comparison between these two gene profiles showed that the gene expression levels were different between BE and CIM epithelia. These 141 genes probably relate to the occurrence and development of BE and the promotion or progression in adenocarcinoma, which might be helpful to understand the molecular mechanism of cell transformation and provides molecular markers and target genes for clinical diagnosis, prevention, treatment of BE and esophageal adenocarcinoma and most of adenocarcinomas at GEJ. The gene expression difference between BE and CIM detected by gene chip might provide a new direction for diagnosis, therapy and prevention of BE.
Edited by Wang XL and Chen WW Proofread by Xu FM
| 1. | O'Connor JB, Falk GW, Richter JE. The incidence of adenocarcinoma and dysplasia in Barrett's esophagus: report on the Cleveland Clinic Barrett's Esophagus Registry. Am J Gastroenterol. 1999;94:2037-2042. [PubMed] |
| 2. | Geboes K. Barrett's esophagus: the metaplasia-dysplasia-carcinoma sequence: morphological aspects. Acta Gastroenterol Belg. 2000;63:13-17. [PubMed] |
| 3. | Morales TG, Sampliner RE, Bhattacharyya A. Intestinal metaplasia of the gastric cardia. Am J Gastroenterol. 1997;92:414-418. [PubMed] |
| 4. | Voutilainen M, Färkkilä M, Juhola M, Mecklin JP, Sipponen P. Complete and incomplete intestinal metaplasia at the oesophagogastric junction: prevalences and associations with endoscopic erosive oesophagitis and gastritis. Gut. 1999;45:644-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Spechler SJ, Zeroogian JM, Antonioli DA, Wang HH, Goyal RK. Prevalence of metaplasia at the gastro-oesophageal junction. Lancet. 1994;344:1533-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 356] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 6. | Goldstein NS, Karim R. Gastric cardia inflammation and intestinal metaplasia: associations with reflux esophagitis and Helicobacter pylori. Mod Pathol. 1999;12:1017-1024. [PubMed] |
| 7. | Pereira AD, Suspiro A, Chaves P, Saraiva A, Glória L, de Almeida JC, Leitão CN, Soares J, Mira FC. Short segments of Barrett's epithelium and intestinal metaplasia in normal appearing oesophagogastric junctions: the same or two different entities. Gut. 1998;42:659-662. [PubMed] |
| 8. | Ruol A, Parenti A, Zaninotto G, Merigliano S, Costantini M, Cagol M, Alfieri R, Bonavina L, Peracchia A, Ancona E. Intestinal metaplasia is the probable common precursor of adenocarcinoma in barrett esophagus and adenocarcinoma of the gastric cardia. Cancer. 2000;88:2520-2528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 9. | Byrne JP, Mathers JM, Parry JM, Attwood SE, Bancewicz J, Woodman CB. Site distribution of oesophagogastric cancer. J Clin Pathol. 2002;55:191-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Hamilton SR, Smith RR, Cameron JL. Prevalence and characteristics of Barrett esophagus in patients with adenocarcinoma of the esophagus or esophagogastric junction. Hum Pathol. 1988;19:942-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 173] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett's esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 12. | Sharma P, Weston AP, Morales T, Topalovski M, Mayo MS, Sampliner RE. Relative risk of dysplasia for patients with intestinal metaplasia in the distal oesophagus and in the gastric cardia. Gut. 2000;46:9-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 127] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 13. | Xu SH, Qian LJ, Mou HZ, Zhu CH, Zhou XM, Liu XL, Chen Y, Bao WY. Difference of gene expression profiles between esophageal carcinoma and its pericancerous epithelium by gene chip. World J Gastroenterol. 2003;9:417-422. [PubMed] |
| 14. | Barrett MT, Yeung KY, Ruzzo WL, Hsu L, Blount PL, Sullivan R, Zarbl H, Delrow J, Rabinovitch PS, Reid BJ. Transcriptional analyses of Barrett's metaplasia and normal upper GI mucosae. Neoplasia. 2002;4:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |