Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2666
Revised: January 13, 2004
Accepted: February 1, 2004
Published online: September 15, 2004
AIM: To establish and assess the methods for quantitative detection of serum duck hepatitis B virus (DHBV) DNA by quantitative membrane hybridization using DHBV DNA probe labeled directly with alkaline phosphatase and fluorescence quantitative PCR (qPCR).
METHODS: Probes of DHBV DNA labeled directly with alkaline phosphatase and chemiluminescent substrate CDP-star were used in this assay. DHBV DNA was detected by autoradiography, and then scanned by DNA dot-blot. In addition, three primers derived from DHBV DNA S gene were designed. Semi-nested primer was labeled by AmpliSensor. Standard curve of the positive standards of DHBV DNA was established after asymmetric preamplification, semi-nested amplification and on-line detection. Results from 100 samples detected separately by alkaline phosphatase direct-labeled DHBV DNA probe with dot-blot hybridization and digoxigenin-labeled DHBV DNA probe hybridization. Seventy samples of duck serum were tested by fluorescent qPCR and digoxigenin-labeled DHBV DNA probe in dot-blot hybridization assay and the correlation of results was analysed.
RESULTS: Sensitivity of alkaline phosphatase direct-labeled DHBV DNA probe was 10 pg. The coincidence was 100% compared with digoxigenin-labeled DHBV DNA probe assay. After 30 cycles, amplification products showed two bands of about 180 bp and 70 bp by 20 g/L agarose gel electrophoresis. Concentration of amplification products was in direct proportion to the initial concentration of positive standards. The detection index was in direct proportion to the quantity of amplification products accumulated in the current cycle. The initial concentration of positive standards was in inverse proportion to the number of cycles needed for enough quantities of amplification products. Correlation coefficient of the results was (0.97, P < 0.01) between fluorescent qPCR and dot-blot hybridization.
CONCLUSION: Alkaline phosphatase direct-labeled DHBV DNA probe in dot-blot hybridization and fluorescent qPCR can be used as valuable means to quantify DHBV DNA in serum.
- Citation: Chen YX, Huang AL, Qi ZY, Guo SH. Establishment and assessment of two methods for quantitative detection of serum duck hepatitis B virus DNA. World J Gastroenterol 2004; 10(18): 2666-2669
- URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2666
Duck hepatitis B virus (DHBV) infection model is an important referenced animal model for studying HBV infection and viral replication and evaluating antiviral agents[1-8]. HBV DNA level in serum is an important evidence for assessing antiviral effect. At present, it is difficult to accurately define the quantity of HBV DNA, though many methods such as competitive PCR, Chiron Quantiplex branched DNA (bDNA) assay, Abbott Genostics solution hybridization assay and Digene Hybrid Capture System have been reported which are not suitable for clinical application because of time- and cost-consuming process. HBV DNA quantitative PCR has been used in clinical detection , but no reports were presented about DHBV DNA quantitative PCR. With the development of non-isotope-labeled nucleic acid probes, especially chemiluminescent-labeled probes[19-21], the probe-sensitivity has been improved obviously. In order to establish a quick, sensitive and specific method for quantitative detection of DHBV DNA in serum, we detected DHBV DNA in serum sample with alkaline phosphatase direct-labeled DHBV DNA probe with dot-blot hybridization and fluorescent qPCR.
DHBV serum was obtained from Institute of Viral Hepatitis, Chongqing University of Medical Sciences, Chongqing, China. pBR325 containing DHBV DNA sequence was extracted, digested with EcoR I and recovered according to the kit protocol. Then quantification was made with DUR Series 600. DNA fragment diluted with water to a series concentrations of 2.76 × 106 to 2.76 × 101 copies/µL was used as positive standard. Reagents and instruments such as alkaline phosphatase direct-labeled kit (Amersham Pharmacia Biotech), Wizard Plus Midipreps DNA purification system (Promega), agarose gel DNA extraction kit (Roche), DUR Series 600 (Beckman), scanning apparatus (Vuego Scan, Brisa-620ST), AmpliSensor kit (Biotronics), Taq DNA polymerase (Roche), AG-9600 Analyzer and Amplicor monitor (Biotronics) were used in the study. Primers derived from DHBV DNA S gene were 5’-TGGCCTAATCGGATTACTGG-3’ (Limited primer, 20 bp), 5’-CCTGGGCATCCCCACGGGCAGG-3’ (Excess primer, 22 bp) and 5’-GGGACGCGCGCTTTCCAAGATACTG-3’ (Semi-nested primer, 25 bp). The three primers were synthesized by Seagon, Shanghai.
Detection of serum DHBV DNA with a membrane hybridization assay using alkaline phosphatase direct-labeled DHBV DNA probe DHBV DNA fragment was diluted to a concentration of 10 ng/µL and labeled referring to alkaline phosphatase direct-labeling protocol. The probe can be stored in 50 mL/L glycerol at -20 °C for up to six months. DHBV DNA in the concentration series (100 ng, 10 ng, 1 ng, 100 pg, 10 pg) was dotted on nitric fibrous membrane in order to determine the probe sensitivity. The specificity was detected with human serum, duck serum and rat serum. The serum sample (40 µL) was dotted on nitric fibrous membrane and 1 mol/L NaOH was used for DNA degeneration. The membrane was baked at 80 °C for 2 h and prehybridized for at least 15 min, then 100 ng of the labeled probe was added to the buffer used for the prehybridization stage to hybridize at 55 °C overnight in a shaking water bath. The membrane was washed with the primary wash buffer for 20 min at 55 °C and with secondary wash buffer for 10 min at room temperature. The excess secondary wash buffer was drained from the blots and the membrane was placed on a clean, flat surface. Detection reagent was piped onto the blots (30-40 µL/cm2) for 2-5 min. The DNA blots were placed side up in the film cassette and a sheet of autoradiography film was placed on top of the blots. The cassette was closed and exposed for 1 h at room temperature. The blots on the film were scanned with scanning apparatus and analysed with Discovery Series Quantity One [BioRad, volume (mm3) = intensity × Area].
Fluorescent qPCR for serum DHBV DNA ligation Five × coupling reagent 10 µL, 5 × AmpliSensor 10 µL, semi-nested primer 0.5 µg, water 30 µL were added to a microcentrifuge tube at room temperature. The tube was incubated for 90 min at 37 °C, then the reaction was terminated by adding 50 µL of deionized water and heating to 90 °C for 5 min to denature the enzymes. The ligated product was labeled as 5 × AmpliSensor primer stock.
Ligation efficiency Two of the sample wells (5 × AmpliSensor primer 1 µL, 1 × PCR buffer 4 µL and PCR extension mixture 5 µL per well), two for negative control and two for standard (5 × AmpliSensor primer 0.1 µL, 1 × PCR buffer 4.9 µL and PCR extension mixture 5 µL per well), two for blank control (water 5 µL, PCR extension mixture 5 µL per well) were designated. The negative control served as a control of baseline signal, whereas the standard was a reference of maximum energy transfer. The reaction mixture was heated to 94 °C for 20 s, 55 °C for 20 s, 72 °C for 30 s, and then 20 °C for 30 s to equilibrate the signal. The fluorescence was read under the coupling mode using AG-9600 Analyzer. Upon finishing, data were saved as baseline reading. One unit of Taq DNA polymerase was added to both the sample and standard wells and the reactions were subjected to three additional thermal cycles. Each cycle consists of 20 s at 94 °C, 20 s at 55 °C and 30 s at 72 °C. At the end of the cycling, reactions were cooled down to 20 °C for 30 s. The fluorescence reading was repeated under the same mode. When the data were saved, the coupling efficiency was displayed on the screen.
Asymmetric preamplification Length of amplification fragment was 182 bp. Ten × PCR buffer 110 µL, 40 mmol/L MgCl2 110 µL, 10 × dNTP 110 µL, 10 × primer 110 µL, 10 × terminal primer 110 µL, H2O 330 µL were added. PCR master mix is intended for 100 reactions. The wells for negative (addition of 5 µL H2O), Apex, standards (addition of 5 µL positive standards), sample (addition of 5 µL sample) were designated. PCR master mix 5 µL was added into each well except the well designated as Apex, to which 10 µL of 1 × PCR buffer instead of 0.5 U Taq DNA polymerase was added. PCR cycle was repeated for 25 times: 20 s at 94 °C, 20 s at 55 °C and 30 s at 72 °C. At the end of the cycle, the reaction was at 72 °C for 30 s and cooled down to 4 °C for 2 min.
Semi-nested PCR and on-line detection A 700-bp fragment was amplified as follows: The 5 × AmpliSensor primer stock 66 µL, 10 × PCR buffer 33 µL, 40 mmol/L MgCl2 33 µL and H2O 198 µL were added into a 0.5 mL microcentrifuge tube to make the AmpliSensor mix. AmpliSensor mix 5 µL was aliquoted to each reaction. PCR cycles were: At 94 °C for 20 s, at 55 °C for 30 s and at 72 °C for 30 s. The reaction was cooled for 30 s at 20 °C to establish a signal equilibrium, and then fluorescence reading was acquired under the standard mode using an AG-9600 Analyzer. Data were saved as a baseline reading. Thermal cycling was resumed for a defined number of cycles. Fluorescence reading was repeated under the same mode, and data were saved as an assay reading. A detection index calculated by APAS software was derived from each reading. The magnitude of the detection index was directly proportional to the quantity of amplification product accumulated at the current cycle. The report window was activated to reveal the quantitative data of each sample.
Assessment of the two methods Alkaline phosphatase direct-labeled DHBV DNA probe with dot-blot hybridization was compared with digoxigenin-labeled DHBV DNA probe with dot-blot hybridization for 100 samples, and then the correlation of 70 samples between fluorescent qPCR and dot-blot hybridization with digoxigenin-labeled probe was analysed.
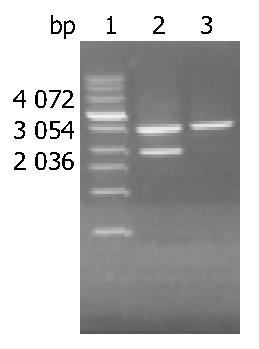
The recovered DHBV DNA fragment after digestion of EcoR I was about 3.0 kb and the concentration was 28 μg/mL (A260/A280 = 1.9) (Figure 1).
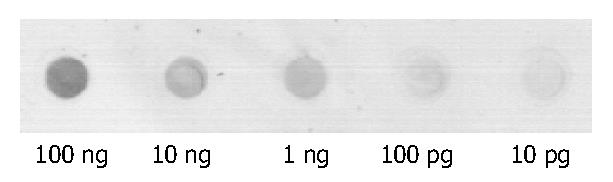
Sensitivity of alkaline phosphatase direct-labeled probe was 10 pg without nonspecific signs (Figure 2, Table 1).
| Genera | Samples (n) | DHBV DNA positive samples (n) | DHBV DNA negative samples (n) |
| Duck serum | 20 | 10 | 10 |
| Human serum | 20 | 0 | 20 |
| Rat serum | 20 | 0 | 20 |
Compared with digoxigenin-labeled DHBV DNA probe, the detection sensitivity of alkaline phosphatase direct-labeled probe was 100% (60/60), the specificity was 100% (40/40) and the coincidence was 100% (80/80) (Table 2).
| Probe | Positive samples (n) | Negative samples (n) |
| Digoxigenin-labeled probe | 60 | 40 |
| Alkaline phosphatase | 60 | 40 |
| direct-labeled probe |
The detection indices of semi-nested PCR and AmpliSensor assay for sample wells were 424.79 and 551.28, respectively. A “+” status was assigned to samples with ligation efficiency greater than 70. The primer could be used in asymmetric preamplification.
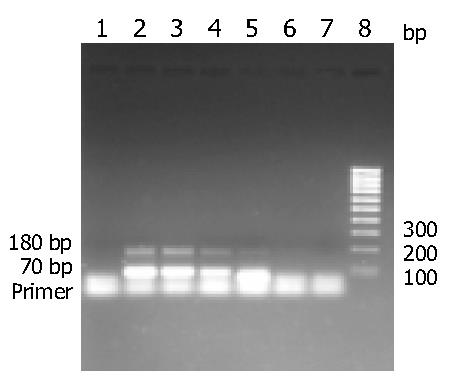
After 30 cycles, amplification products showed two bands of about 180 bp and 70 bp by 2% agarose gel electrophoresis. The concentration of amplification products was in direct proportion to the initial concentration of the positive standards (Figure 3).
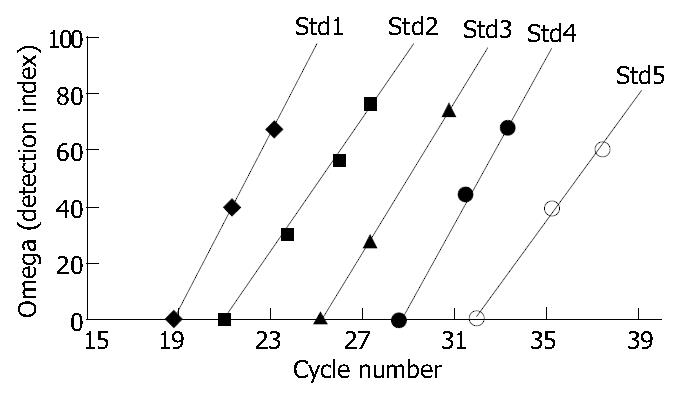
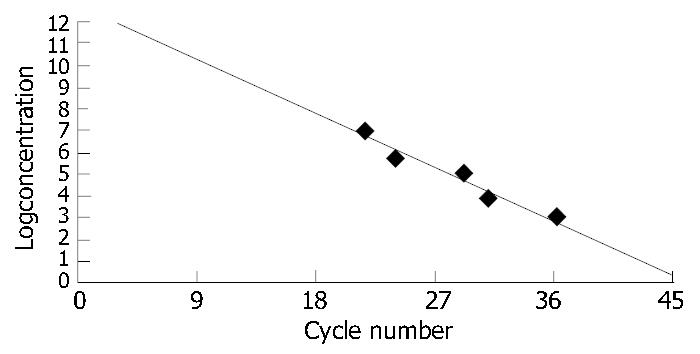
The detection index curves Std1-Std5 showed that the detection index was in direct proportion to the quantities of amplification products at the current cycle (Figure 4). The concentration curve showed that the initial concentration of the positive standards was in inverse proportion to the number of cycles needed for enough quantities of amplification products and the correlation coefficient between initial concentration and number of cycles was 0.992 (Figure 5).
Seventy serum samples of ducks were tested by fluorescent qPCR and digoxigenin-labeled DHBV DNA probe with dot-blot hybridization, and the correlation of results was analysed. Correlation coefficient of the results was 0.97 (P < 0.01) between the two methods. DHBV DNA could not be found by dot-blot hybridization, but it could be detected by qPCR when DHBV DNA loads < 102 copies/mL (Table 3).
| Serum samples (n) | qPCR (mean ± SD) (copy/mL) | Dot-blot hybridization using digoxigenin-labeled probe (mean ± SD) (volume, mm3) |
| 10 | (2.37 ± 1.70) × 102 | - |
| 10 | (2.44 ± 1.71) × 103 | 1155 ± 25 |
| 10 | (4.47 ± 1.96) × 104 | 1241 ± 32 |
| 10 | (5.02 ± 2.14) × 105 | 1353 ± 27 |
| 10 | (1.83 ± 1.03) × 106 | 1428 ± 25 |
| 10 | (4.81 ± 2.61) × 107 | 1550 ± 40 |
| 10 | (5.16 ± 2.72) × 108 | 1748 ± 118 |
Though probes labeled with radioisotope are still used to detect nucleic acids because of its high sensitivity and complete operation rule established many years ago, the safety and halflife of radioisotope must be taken into account. At present, labeling and detecting with non-radioisotope have made significant progress, and the sensitivity of the probe labeled with chemiluminescence has reached 50 fg. Two methods of labeling with non-radioisotope were available in the present study: non-direct-labeling such as random primed DNA-labeling with digoxigenin-dUTP[22] and direct-labeling such as DNA-labeling with alkaline phosphatase[23]. DNA-labeling with alkaline phosphatase is a new method with advantages of quickness, convenience and high sensitivity. Some procedures such as long time incubation with antibody and blocking reagent and elution can be omitted. The probe can be detached from hybridizing membrane and therefore the membrane can be used to hybridize with another probe sensitively and conveniently. Furthermore, alkaline phosphatase may be detected with chemiluminescence (dioxetane) and autoradiography or photoscanning apparatus. Chen et al[21] reported that the sensitivity of dot-blot hybridization with Epstein-Barr virus probe coupled with alkaline phosphatase was 4 pg, which is similar to our results that the sensitivity of dot-blot hybridization with DHBV probe labeled by alkaline phosphatase was 10 pg. It is necessary to optimize labeling method of alkaline phosphatase for improving the sensitivity of the probe. This study showed that the alkaline phosphatase direct-labeled probe could be used to evaluate antiviral activity in DHBV infection animal model.
AmpliSensor-PCR is a real-time, quantitative tool for PCR-based detection. The assay is based on the principle that fluorescence resonance energy transfer can be used to detect duplex formation between complementary nucleic acid strands. If the two complementary strands are labeled with donor and acceptor fluorophores, respectively, fluorescence resonance energy transfer between the fluorophores is facilitated when the strands are base-paired, or eliminated when the base-pair is disrupted. In this way, the extent of energy transfer can be used to measure the amount of duplex formation between the fluorophore-labeled oligonucleotide duplexes, thereby the extent of duplex formation mediated DNA polymerase. The measured fluorescence intensity is in proportion to the quantity of AmpliSensor duplex left at the end of each amplification cycle. The decrease in the fluorescence intensity correlates proportionately to the initial target dosage and the extent of amplification. The signal-generating mechanism of AmpliSensenor assay is tightly linked to the priming event. However, unlike typical PCR, where single-stranded primers are constantly subject to the possibility of nonspecific priming, the AmpliSensor primer is sequestered in a double-stranded form and stabilized against random priming. The AmpliSensor is a quasi-stable signal duplex of two oligonucleotides each labeled with an energy donor and acceptor fluorophore, respectively. Two strands of the AmpliSensor are unequal in length with the long strand 5’ overhanging the short one by 7 nucleotides (5’-GCGTCCC-3’). For effective ligation, the semi-nested primer should encompass at its 5’ end a “hook” sequence, that is, 5’-GGGACGC-3’, complementing the overhang of the AmpliSensor. The fluorescence signal correlates to the overall energy transfer efficiency in a predictable, sequence-specific manner, and the amplified product can be monitored directly. As a simple, homogeneous assay for PCR quantification, AmpliSensor technology is target-specific, and it is amenable to the normalization of signal loading and sample loading. Thus it can reliablly count the amplified product while it accounts for the efficiency difference among reactions.
At present, fluorescence quantitative PCR has been applied in the fields of clinical diagnosis for hepatitis B and C, evaluating and monitoring of antiviral effect, prediction of interferon effect, etc.[24,25].
DHBV and HBV are two members of the hepadnavirus family of viruses. They both display similar molecular biological character and 40% nucleic acid sequence homology. We usually make use of DHBV model to learn human hepatitis B about its infection and replication strategies or antiviral effect and to screen new anti-HBV drugs. In previous studies, dot-blot hybridization, Southern blotting and in situ hybridization with digoxigenin-labeled probe were used to detect DHBV DNA in serum or liver, but the advanced AmpliSensor-PCR and a membrane hybridization assay with alkaline phosphatase direct-labeled DHBV DNA probe could provide more objective, more valuable experimental data for developing new anti-HBV drugs and elucidating pathogenesis of hepatitis B.
Edited by Chen WW Proofread by Xu FM and Zhu LH
| 1. | Foster WK, Miller DS, Marion PL, Colonno RJ, Kotlarski I, Jilbert AR. Entecavir therapy combined with DNA vaccination for persistent duck hepatitis B virus infection. Antimicrob Agents Chemother. 2003;47:2624-2635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 2. | Cooper A, Paran N, Shaul Y. The earliest steps in hepatitis B virus infection. Biochim Biophys Acta. 2003;1614:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Dong B, Shao XW, Tao PZ. [Inhibition of duck hepatitis B virus DNA replication by antisense phosphorothioate oligodeoxynucleotides in vitro and in vivo]. Zhonghua Shiyan He Linchuangbingduxue Zazhi. 2003;17:25-27. [PubMed] |
| 4. | Seignères B, Martin P, Werle B, Schorr O, Jamard C, Rimsky L, Trépo C, Zoulim F. Effects of pyrimidine and purine analog combinations in the duck hepatitis B virus infection model. Antimicrob Agents Chemother. 2003;47:1842-1852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Le Guerhier F, Thermet A, Guerret S, Chevallier M, Jamard C, Gibbs CS, Trépo C, Cova L, Zoulim F. Antiviral effect of adefovir in combination with a DNA vaccine in the duck hepatitis B virus infection model. J Hepatol. 2003;38:328-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Zoulim F, Berthillon P, Guerhier FL, Seigneres B, Germon S, Pichoud C, Cheng YC, Trepo C. Animal models for the study of HBV infection and the evaluation of new anti-HBV strategies. J Gastroenterol Hepatol. 2002;17 Suppl:S460-S463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Thermet A, Rollier C, Zoulim F, Trepo C, Cova L. Progress in DNA vaccine for prophylaxis and therapy of hepatitis B. Vaccine. 2003;21:659-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Lian Y, Tan J, Deng Z. [Experimental study of shuangcao granule no. 1 on duck hepatitis B virus in ducklings]. Zhongguo Zhongxiyi Jiehe Zazhi. 2000;20:530-532. [PubMed] |
| 9. | Sidorkiewicz M, Bzorska A, Józwiak B, Jabłkowski M, Szemraj J, Lewandowska U. A competitor DNA template for the molecular quantification of the hepatitis B virus. Cell Mol Biol Lett. 2003;8:799-808. [PubMed] |
| 10. | Drosten C, Weber M, Seifried E, Roth WK. Evaluation of a new PCR assay with competitive internal control sequence for blood donor screening. Transfusion. 2000;40:718-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Pawlotsky JM, Bastie A, Hézode C, Lonjon I, Darthuy F, Rémiré J, Dhumeaux D. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J Virol Methods. 2000;85:11-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 57] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Gilbert N, Corden S, Ijaz S, Grant PR, Tedder RS, Boxall EH. Comparison of commercial assays for the quantification of HBV DNA load in health care workers: calibration differences. J Virol Methods. 2002;100:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Jardi R, Rodriguez F, Buti M, Costa X, Cotrina M, Valdes A, Galimany R, Esteban R, Guardia J. Quantitative detection of hepatitis B virus DNA in serum by a new rapid real-time fluorescence PCR assay. J Viral Hepat. 2001;8:465-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Hwang SJ, Lu RH, Wood ML, Wang YJ, Chang FY, Lee SD. Comparison of the nucleic acid-based crosslinking hybridization assay and the branched DNA signal amplification assay in the quantitative measurement of serum hepatitis B virus DNA. J Clin Lab Anal. 1999;13:296-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Kao JH, Wood M, Chen PJ, Lai MY, Chen DS. Comparison of two methods for quantification of hepatitis B viral DNA. J Gastroenterol Hepatol. 1999;14:423-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Nagata I, Colucci G, Gregorio GV, Cheeseman P, Williams R, Mieli-Vergani G, Vergani D. The role of HBV DNA quantitative PCR in monitoring the response to interferon treatment in chronic hepatitis B virus infection. J Hepatol. 1999;30:965-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Kessler HH, Preininger S, Stelzl E, Daghofer E, Santner BI, Marth E, Lackner H, Stauber RE. Identification of different states of hepatitis B virus infection with a quantitative PCR assay. Clin Diagn Lab Immunol. 2000;7:298-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Chen T, Luk JM, Cheung ST, Yu WC, Fan ST. Evaluation of quantitative PCR and branched-chain DNA assay for detection of hepatitis B virus DNA in sera from hepatocellular carcinoma and liver transplant patients. J Clin Microbiol. 2000;38:1977-1980. [PubMed] |
| 19. | Kricka LJ. Stains, labels and detection strategies for nucleic acids assays. Ann Clin Biochem. 2002;39:114-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 90] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Sander T, Olson S, Hall J, Siebert M, Grooms K, Heisler L, de Arruda M, Neri B. Comparison of detection platforms and post-polymerase chain reaction DNA purification methods for use in conjunction with Cleavase fragment length polymorphism analysis. Electrophoresis. 1999;20:1131-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Chen Y, Chen Y, Lii H, Lu Z. Chemiluminescence detection of epstein-barr virus DNA with an oligonucleotide probe. Clin Chim Acta. 2000;298:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 22. | Ying C, Van Pelt J, Yap SH, De Clercq E, Neyts J. Use of digoxigenin-labelled probes for the quantitation of HBV-DNA in antiviral drug evaluation. J Virol Methods. 1999;81:155-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Chen Y, Huang A, Qi Z, Shan Y, Sun H. [Establishment and evaluation of the method for detecting HBV DNA in serum using HBV DNA probe labeled directly by alkaline phosphatase]. Zhonghua Ganzangbing Zazhi. 2002;10:429-431. [PubMed] |
| 24. | Zhou X, Gao Z, Yao J. [Relationship between HBV DNA serum level and acute exacerbation of the disease in chronic hepatitis B patients]. Zhonghua Shiyan He Linchuangbingduxue Zazhi. 1999;13:335-339. [PubMed] |
| 25. | Nie Q, Wang P, Zhou Y. [Quantitation of hepatitis C virus RNA in amniotic fluid of gravida infected by hepatitis C virus]. Zhonghua Fuchanke Zazhi. 2002;37:19-21. [PubMed] |