Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2130
Revised: February 26, 2004
Accepted: March 2, 2004
Published online: July 15, 2004
AIM: To investigate the multiple gene differential expression patterns in human ischemic liver and to produce the evidence about the hepatic ischemic safety time.
METHODS: The responses of cells to hepatic ischemia and hypoxia at hepatic ischemia were analyzed by cDNA microarrary representing 4000 different human genes containing 200 apoptotic correlative genes.
RESULTS: There were lower or normal expression levels of apoptotic correlative genes during the periods of hepatic ischemia for 0-15 min, the maintenance homostatic genes were expressed significantly higher at the same time. But at the hepatic ischemia for 30 min, the expression levels of maintenance homeostatic genes were down-regulated, the expressions of many apoptotic correlative genes and nuclear transcription factors were activated and up-regulated.
CONCLUSION: HIF-1, APAF-1, PCDC10, FBX5, DFF40, DFFA XIAP, survivin may be regarded as the signal genes to judge the degree of hepatic ischemic-hypoxic injure, and the apoptotic liver cell injury due to ischemia in different time limits. The safe limit of human hepatic warm ischemic time appears to be generally less then 30 min.
- Citation: Lu QP, Cao TJ, Zhang ZY, Liu W. Multiple gene differential expression patterns in human ischemic liver: Safe limit of warm ischemic time. World J Gastroenterol 2004; 10(14): 2130-2133
- URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2130.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2130
The safety limit of human hepatic warm ischemic time is closely correlated with the recovery of hepatic function and the patient’s prognosis after liver transplantation, serious hepatic injury and hepatectomy[1-7]. Owing to the development in hepatic surgical operation techniques and perioperative period, numerous experts discovered that the safety limit of human hepatic warm ischemic time could be prolonged further. But the critical problem of what degree the safety limit should be generally controlled is drawing special attention of the whole hepatic surgery. The cell apoptosis we have found now is one of the main cell death forms influencing visceral functions after the liver got ischemic injury[8-12]. But because too many genes are involved in cell apoptosis, the expressions in differential ischemic safety time of the main controlling gene influencing the safety limit of human hepatic warm ischemic time remain unclear. For such a reason, by cDNA microarray, we investigated the multiple genes differential expression patterns in human ischemic liver for the purpose of producing some scientific evidence for the hepatic ischemic safety time.
All the tissue specimens in question came from the hepatic tissues contributed by the unconscious patients dying of external brain injury whose other organic functions were normal (Table 1). Each group consisted of 5 specimens. The specimens were immediately stored in liquid N2 after resection. All contributors had no hepatic disease, and the liver function remains normal before resection. The liver tissue construction was proved normal by pathologic examinations.
| Group | Experiment group | Control group |
| Group 1 | 15 min ischemic tissue | Normal liver tissue |
| Group 2 | 30 min ischemic tissue | Normal liver tissue |
| Group 3 | 30 min ischemic tissue | 15 min ischemic tissue |
TriZol and Script II reverse transcriptases were bought from American Life Technologies, Ltd. cDNA microarray containing 200 apoptotic correlative genes and biochip hybridization kit were from Shanghai Biotstar Gene chip, Ltd.
Total RNA was extracted from human liver tissues in each group following the single step extraction (see details in the manual).
An equal volume of RNA was extracted from each group and 60 μg RNA was added into 50 μL reverse transcription system. The experiment group was labeled with Cy5-d CTP and the control group with Cy5-d CTP (see the way of labeling in the manual of the test kit). The chip was denatured and prehybridized for one time in advance. After denatured at 95 °C for 5 min, the probe mixture was added on the microarray and covered with a hyi slip. The slide was then placed in a constant temperature hybridization chamber for hybridization at 42 °C for 20 h. After hybridization, slides were washed 10 min in each of 2 × SSC with 2 g/L GSDS, 0.1 × SSC with 2 g/L GSDS, 0.1 × SSC, then dried at room temperature. The hybridization in each group was replicated 3 times.
The chip was scanned by GenePix 4000B laser scanner at 2 wavelengths. The acquired image was analyzed by GenePix Pro 3.0 software. The intensity of each spot at the 2 wavelengths cut off the additional background signal represented the quantity of Cy3-dUTP and Cy5-dUTP respectively. Each ratio value of Cy3 to Cy5 was computed. The 2 overall intensities were normalized by a coefficient according to the ratio of the located 88 housekeeping genes, 43 negative control genes, 45 positive control genes and 3 blank locations. We defined two standards to screen out each differentially expressed gene. The ratio value > 2 represented the level of the gene expression elevated significantly, the ratio value < 0.5 represented the level of the significantly depressed gene expression. The data about the genes whose ratio value > 2 or < 0.5 were screened out in the comparisons of normal control group with 15 min ischemic group or 30 min ischemic group, then the variation of the ratio value was observed in the comparison of 15 min ischemic group with 30 min ischemic group and each “± 0.5” change in the ratio value was set as a “+”. All the data were input in the datasheet, and the biological functions of the target genes were analyzed.
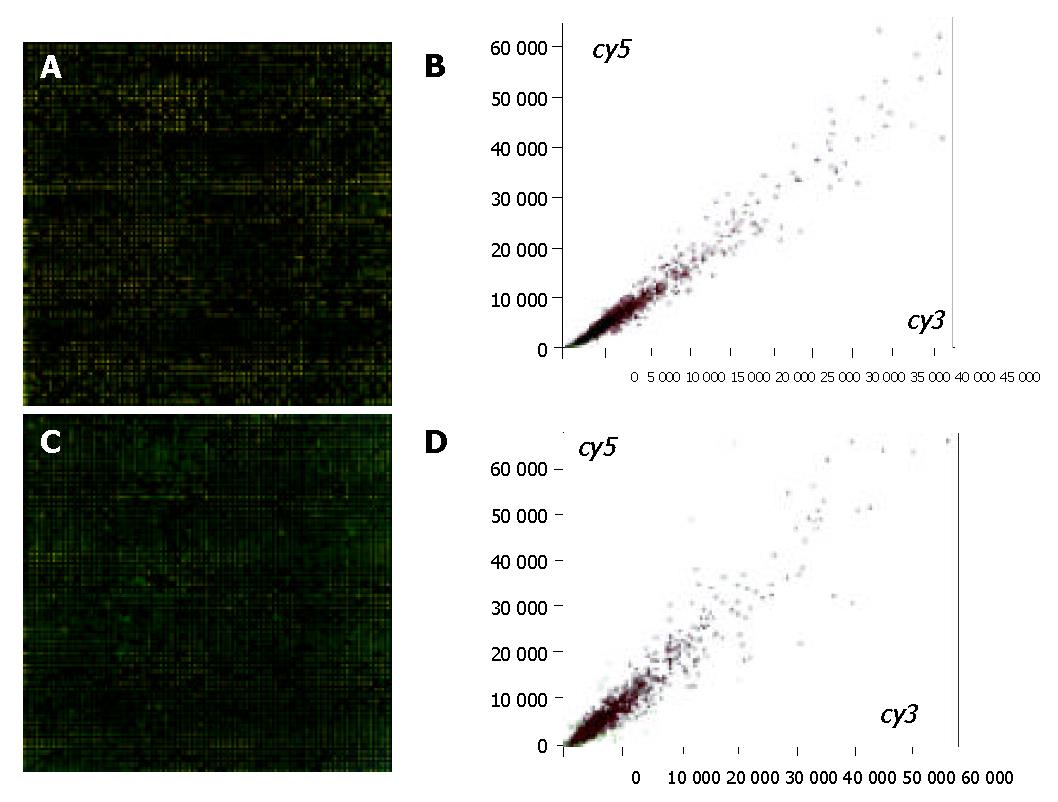
Figure 1 (A and B) shows that the double-colored fluorescent labels in 15 min ischemic group and normal control group were overlapping. A total of 41 differentially expressed genes were identified by cDNA chip between the 15 min ischemic group and normal control group. Among them the ratio values of 33 genes were < 0.5, consisting of 22 genes correlated directly with cell apoptosis regulation, 9 congenic genes of stress-response protein relevant to cell apoptosis, and 3 genes correlated to metabolism regulation, 1 correlated to cell circle regulation, 1 correlated to the cell interpretation and synthesis. In the meantime, there were 8 differential expression genes whose ratio values were > 2, mainly about P43 = mitochondrial elongation factor homolog (GenBank-ID: S75463), human ribosomal protein S14 gene, complete cds (GenBank-ID: humrps 14), H. sapiens mRNA for squalene synthase (GenBank-ID: hssqusyn), human sodium/potassium ATPase beta-2 subunit (atpb2) mRNA, complete cds (GenBank-ID: humatpbii), H. sapiens mRNA for H+-ATP synthase subunit b (GenBank-ID: hsatpsyn), and no gene relevant to apoptosis was highly expressed.
Figure 1 (C and D) shows that the double-colored fluorescent labels in 30 min ischemic group and normal control group were overlapping. A total of 9 differentially expressed genes were identified by cDNA chip between the 30 min ischemic group and normal control group. The ratio values of 3 genes of which were < 0.5, consisting of 2 genes correlated directly with cell apoptosis regulation (the congenic genes of survivin, AB028869 and X-linked inhibitor apoptosis protein, u45880 respectively), and 1 congenic gene (GenBank-ID: af004711) of TPKC1. In the meantime, there were 6 differential expression genes whose ratio values were > 2, of which 3 were the regulation genes correlated directly with cell apoptosis: APAF1 (GenBank-ID: NM-001160), PDCD10 (GenBank-ID: NM-007210) and an unnamed gene (GenBank-ID: AL031714), 1 was the congenic gene (GenBank-ID: AF050127) of HIF1, the functions of last 2 genes’ were not clear.
In the 200 genes, 35 genes were not expressed at the two time limits. The expressions of 167 genes in 30 min ischemic group differed from in 15 min ischemic group. One hundred and nineteen genes were expressed following the rising tendency, of which ratio values of 7 genes were > “+++”, 19 > “++”, 39 > “+”, and 55 genes only expressed the rising tendency but their ratio values were ≤“+”. There are 46 genes following the descending tendency, of which ratio values of 4 genes were > “+++”, 5 > “++”, 12 > “+”, and 25 genes only expressed the descending tendency but their ratio values were ≤“+”. In addition, 16 genes were expressed in 30 min ischemic group, but not in 15 min ischemic group, of which 11 genes were correlated directly with cell apoptosis regulation, 5 genes were expressed in 15 min ischemic group, but not in 30 min ischemic group, of which only 1 gene was correlated directly with cell apoptosis regulation, namely XIAP (GenBank-ID: u45880).
Our data suggested that the apoptosis regulation gene at the hepatic ischemia for 0-30 min presented following features.
In hepatic ischemia for 0-15 min, the apoptosis regulation gene was expressed low or normally. The differentially expressed genes whose ratio values were > 2 (namely, they were expressed following the rising tendency) were mainly the maintenance harmonious genes, such as genes regulate P43 = mitochondrial elongation factor homolog (GenBank-ID: S75463), Human ribosomal protein S14 gene (GenBank-ID: humrps), H. sapiens mRNA for squalene synthase (GenBank-ID: hssqusyn), Na+/K+-ATPase activity α subunit (GenBank-ID: hsatpar) and β subunit(GenBank-ID: humatpbii), H+-ATPase activity (GenBank-ID: hsatpsyn). But they were not highly expressed with the congenic genes correlated directly with cell apoptosis regulation.
In hepatic ischemia for 15-30 min, the expression of apoptosis regulation genes changed greatly. One hundred and nineteen genes were expressed following the rising tendency, of which 59 genes were relevant to the apoptosis regulation. Although some anti-apoptosis genes [such as the congenic gene of bcl-2 (GenBank-ID: AJ006288), the congenic gene of inhibitor apoptosis protein IEX-IL (GenBank-ID: AF071596)] which were expressed the low level, the majority of the higher expressed genes were the apoptotic genes, for example, the congenic gene of apoptotic protease activating factor-1 (APAF-1, GenBank-ID: NM-001160); Homo sapiens F-box protein 5, FBX5 (GenBank-ID: NM-012177); DNA fragmentation factor, 40 ku, subunit, DFF40 (GenBank-ID: AF064019); DNA fragmentation factor, 45 ku, alpha subunit, DFFA (GenBank-ID: NM-004401); p53-induced protein; PIG11(GenBank-ID: NM-006034). Especially, APAF-1 was the only known human homologue of CED-4 and the nuclear element in the formation of apoptosis body[13-16]. Now it is thought as the key element regulating cell apoptosis, and the ratio value of its expression in 30 min hepatic ischemia was an additional “+++” than that in 15 min hepatic ischemia. Meanwhile the ratio value of the congenic gene of PDCD10 (GenBank-ID: NM-007217) was also “++”. The significant rising of expression of the apoptotic genes showed that the unavoidable cell death after serious ischemic injury was a key expression of the serious cell injure. In 15 min hepatic ischemia, the congenic gene of the important X-linked inhibitor apoptosis protein, XIAP (GenBank-ID: u45880) changed from the normal expression to non-expression, following the obvious descending tendency. Compared with the normal group, the expression of the survivin congenic gene (GenBank-ID: AB028869) descended greatly with the ratio value < 0.5. The codogenic genes (GenBank-ID: S75463, humrps, hssqusyn, hsatpar, humatpbii, hsatpsyn and so on) relevant to the internal circumstance and the organelle functions were expressed obviously in the period of 0-15 min hepatic ischemia, but in 15-30 min period their expression descended to “+”-“++”. The congenic gene of human hypoxiainducible factor 1 HIF-1 alpha gene (GenBank-ID: AF050127) was expressed apparently (increased to “+++”). Lots of the regulation genes of nucleus translation factors were expressed notably for example, the codogenic gene of EIF4G2 (eukaryotic translation-induction factor 4 gamma, 2 (EIF4G2) mRNA, GenBank-ID: NM-001418), NF-κB family, TNF receptor associated factor 6 (TRAF6) responsible for the existence and death of cells (GenBank-ID: hsu78798), increased to “++”. The latter was a transmission factor participating in IL-1 signal transmission and activating the signal transmission about apoptosis of the nuclear factor NF-κB[17-20].
The data suggested that in 0-15 min hepatic ischemia, the gene regulation model inside of the cells was likely to better and maintain the harmonious of the internal circumstance to keep the wholeness of cell and organ functions even by controlling ion channel and organelle functions. At the same time, although acute ischemic-hypoxic injury changed the structure and functions of cells, the expression of each kind of genes relevant to apoptosis, according to the analysis of the level of gene regulation, remained on the low level, that is to say, the occurrence of apoptosis was not a main direction yet. With the time of ischemia and hypoxia passed, when hepatic ischemia lasted for 30 min, acute ischemic-hypoxic injury perhaps would lead to the significant rising of HIF-1, and the cell gene expression model responsible for the existence or death of the cells also was changed completely. The expressions of all kinds of regulation genes maintaining cells and the internal cellular circumstance descended obviously, while NF-κB and many genes relevant to apoptosis were activated evidently and notably expressed. Therefore, it is suggested that cell death is unavoidable when hepatic ischemia lasts for 30 min. It is true that some experts deduced that the time of the human hepatic ischemia could be prolonged beyond the safe limit according to clinical experience in the progress after liver operation. Yet when dynamic changes of gene expressions after liver suffered ischemic injury are analyzed, it is suggested that the safe limit of one time hepatic ischemia should be less than 30 min, namely, when key death genes such as APAF-1, PDCD10, FBX5, DFF40, DFFA are activated evidently and expressed significantly, and before the obvious descending of inhibitor apoptosis genes such as XIAP, survivin. In the meantime it is suggested that HIF-1, APAF-1, PCDC10, XIAP, survivin may be regarded as the signal gene to judge the degree of hepatic ischemic-hypoxic injury, and the apoptotic liver cell injury due to ischemia in different time limits.
Edited by Wang XL Proofread by Chen WW and Xu FM
| 1. | Makuuchi M, Mori T, Gunvén P, Yamazaki S, Hasegawa H. Safety of hemihepatic vascular occlusion during resection of the liver. Surg Gynecol Obstet. 1987;164:155-158. [PubMed] |
| 2. | Mori T, Makuuchi M, Kobayashi J, Sukigara M, Yamasaki S, Hasegawa H. [Clinical studies on changes in serum transaminase, lactate dehydrogenase, total bilirubin and alkaline phosphatase levels after hepatectomy with and without the hemihepatic vascular occlusion technique]. Nihon Geka Gakkai Zasshi. 1985;86:837-845. [PubMed] |
| 3. | Gotoh M, Monden M, Sakon M, Kanai T, Umeshita K, Nagano H, Mori T. Hilar lobar vascular occlusion for hepatic resection. J Am Coll Surg. 1994;178:6-10. [PubMed] |
| 4. | Wu CC, Yeh DC, Ho WM, Yu CL, Cheng SB, Liu TJ, P'eng FK. Occlusion of hepatic blood inflow for complex central liver resections in cirrhotic patients: a randomized comparison of hemihepatic and total hepatic occlusion techniques. Arch Surg. 2002;137:1369-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Liu DL, Jeppsson B, Hakansson CH, Odselius R. Multiple-system organ damage resulting from prolonged hepatic inflow interruption. Arch Surg. 1996;131:442-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Emond J, Wachs ME, Renz JF, Kelley S, Harris H, Roberts JP, Ascher NL, Lim RC. Total vascular exclusion for major hepatectomy in patients with abnormal liver parenchyma. Arch Surg. 1995;130:824-830; discussion 830-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Scheele J. [Anatomical and atypical liver resections]. Chirurg. 2001;72:113-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Yin XM, Ding WX. Death receptor activation-induced hepatocyte apoptosis and liver injury. Curr Mol Med. 2003;3:491-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 115] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 9. | Jaeschke H, Lemasters JJ. Apoptosis versus oncotic necrosis in hepatic ischemia/reperfusion injury. Gastroenterology. 2003;125:1246-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 441] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 10. | Sass G, Soares MC, Yamashita K, Seyfried S, Zimmermann WH, Eschenhagen T, Kaczmarek E, Ritter T, Volk HD, Tiegs G. Heme oxygenase-1 and its reaction product, carbon monoxide, prevent inflammation-related apoptotic liver damage in mice. Hepatology. 2003;38:909-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Berberat PO, Katori M, Kaczmarek E, Anselmo D, Lassman C, Ke B, Shen X, Busuttil RW, Yamashita K, Csizmadia E. Heavy chain ferritin acts as an antiapoptotic gene that protects livers from ischemia reperfusion injury. FASEB J. 2003;17:1724-1726. [PubMed] |
| 12. | Topaloglu S, Abbasoglu O, Ayhan A, Sokmensuer C, Kilinc K. Antiapoptotic and protective effects of roscovitine on ischemia-reperfusion injury of the rat liver. Liver Int. 2003;23:300-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2262] [Cited by in RCA: 2264] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 14. | Pan G, O'Rourke K, Dixit VM. Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex. J Biol Chem. 1998;273:5841-5845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 377] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 15. | Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 377] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 16. | Hu Y, Benedict MA, Wu D, Inohara N, Núñez G. Bcl-XL interacts with Apaf-1 and inhibits Apaf-1-dependent caspase-9 activation. Proc Natl Acad Sci USA. 1998;95:4386-4391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 399] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 17. | Matsumura T, Degawa T, Takii T, Hayashi H, Okamoto T, Inoue J, Onozaki K. TRAF6-NF-kappaB pathway is essential for interleukin-1-induced TLR2 expression and its functional response to TLR2 ligand in murine hepatocytes. Immunology. 2003;109:127-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Takatsuna H, Kato H, Gohda J, Akiyama T, Moriya A, Okamoto Y, Yamagata Y, Otsuka M, Umezawa K, Semba K. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J Biol Chem. 2003;278:12144-12150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Kanamori M, Kai C, Hayashizaki Y, Suzuki H. NF-kappaB activator Act1 associates with IL-1/Toll pathway adaptor molecule TRAF6. FEBS Lett. 2002;532:241-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |