Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2072
Revised: January 13, 2004
Accepted: February 1, 2004
Published online: July 15, 2004
AIM: To investigate whether P28 derived from C3d can enhance the immune response to HBV-preS2/S induced by directly injection of naked plasmids containing variable repeats of P28 and HBV-preS2/S in fusion form.
METHODS: One to four copies of C3d-P28 coding gene, amplified by PCR and modified by restriction endonucleases digestion, were subcloned into a eukaryotic expression vector pVAON33 to construct pVAON33-P28, pVAON33-P28.2, pVAON33-P28.3 and pVAON33-P28.4 (pVAON33-P28.[1-4]). HBV-preS2/S coding sequence was then introduced into the pVAON33-P28.[1-4] and identified by both PCR and DNA sequencing. BALB/c mice were primed by intramuscular gene immunization with 100 μg different recombinant plasmids on day 0 and were boosted by subcutaneous inoculation with HBsAg protein (1 μg) 12 wk post-priming. The levels and avidity of specific IgG in sera collected at the indicated times from each group were determined by ELISA and NaSCN-displacement ELISA, respectively.
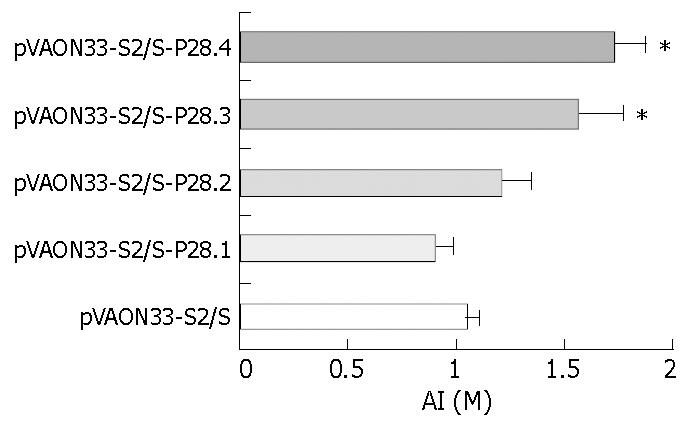
RESULTS: HBsAg specific antibody response was elicited in groups primed with plasmids pVAON33-S2/S-P28.[1-4] and pVAON33-S2/S. However, the response against HBsAg in the groups primed with pVAON33-S2/S-P28.[1-4] was significantly higher than that in pVAON33-S2/S group, the highest level of the specific antibody response was observed in the groups primed with pVAON33-S2/S-P28.4 (P < 0.01). After secondary immunization with specific antigen, the acceleration of antibody levels was significantly higher and faster in the mice primed with DNA expressing preS2/S-P28 fusions than that with DNA expressing preS2/S only (P < 0.05). Interestingly, mice primed with DNA expressing preS2/S-P28.4 fusions maintained the highest levels of anti-HBs antibodies in all animals. The avidity assay showed that the avidity index (AI) collected at 18 wk from mice primed with pVAON33-S2/S-P28.3 and pVAON33-S2/S-P28.4 were significantly higher than that from preS2/S-DNA vaccinated mice (P < 0.01).
CONCLUSION: Different repeats of C3d-P28 can enhance both humoral immune response and avidity maturation of specific antibodies induced by gene immunization, in which four copies of C3d-P28 may be necessary to achieve the most modest antibody response.
- Citation: Wang LX, Xu W, Guan QD, Chu YW, Wang Y, Xiong SD. Contribution of C3d-P28 repeats to enhancement of immune responses against HBV-preS2/S induced by gene immunization. World J Gastroenterol 2004; 10(14): 2072-2077
- URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2072.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2072
The third complement protein (C3) plays a major role in the complement activation pathway, the critical role of the cleavage fragments of C3 in the humoral immune response for both T-dependent and T-independent antigens was reported in studies performed over a quarter of a century ago[1,2]. Complement’s potential use as an adjuvant in vaccines was first suggested when Dempsey et al[2] demonstrated that mice, vaccinated with a genetically engineered construct containing three copies of mouse C3d fused to a model antigen, hen egg lysozyme, increased the efficiency of immunizations by more than 1000-fold. Subsequent studies by Test et al[3] showed that covalent conjugates of C3d and the capsular polysaccharide of serotype 14 streptococcus pneumoniae (PPS14) elicited higher titers to PPS14 in mice than PPS14 only, and furthermore induced a class switch in anti-PPS14 from predominantly IgM to IgG1, which extended the adjuvant effects of C3d to T-independent antigen as well as T-dependent antigen. Recently, studies have further shown that gene immunization with C3d is an effective molecular adjuvant for inducing antibody responses to a range of viral pathogens, including influenza virus[4,5], human immunodeficiency virus[6], and measles virus[7]. The mechanism, by which C3d increases antibody responses, has been hypothesized to reflect the binding of C3d to cluster of differentiation 21 (CD21) on the surface of B-cells or follicular dendritic cells (FDC)[4,5,8,9].
The complement receptor 2 (CR2)-binding site on C3d was located, using chemical fragmentation and peptide mapping studies, between residues 1199 and 1210 of the complement C3 sequence (mature C3 numbering)[10]. A synthetic peptide corresponding to the CR2-binding site on C3d, P28 (C3K1,187-A1,214: 1187KFLTTAKDKNRWEDPGKQLYNVEATSYA1214), as well as other C3d homologous, specifically binds to CR2 expressed on B cell lines, such as Raji cells[10,11]. Binding of P28 stimulates the proliferation of peripheral resting B lymphocytes or CR2-positive B cell lines[11-13]. In addition, the binding of CR2 to P28 peptides and the proliferative response of B cells by these peptides were dose dependent and could be inhibited by soluble C3d or anti-CR2 mAb[12,14]. Furthermore, when a P16 peptide (equivalent to residues 1195-1210 of C3) was coupled to anti-idiotype antibody, it induced a strong idiotype and antigen-specific response in mice[15].
In present study, we selected this active C3d-P28 peptide as a molecular adjuvant according to its binding-ability to CR2 on the B-cells or FDC[10-13,15], and HBV-preS2/S as a model antigen, to seek an enhanced anti-preS2/S antibody response following vaccination with DNA expressing fusions of HBV-preS2/S to variable copies of C3d-P28. Our results showed that immunizations with the preS2/S-P28 fusions DNA not only induced higher primary humoral responses as well as a faster and stronger memory reaction, but also accelerated the avidity maturation of anti-HBs compared with that resulting from immunization with preS2/S only. Our findings argue that four or more repeats of C3d-P28 may be necessary for efficient enhancement of antigen-specific immune responses.
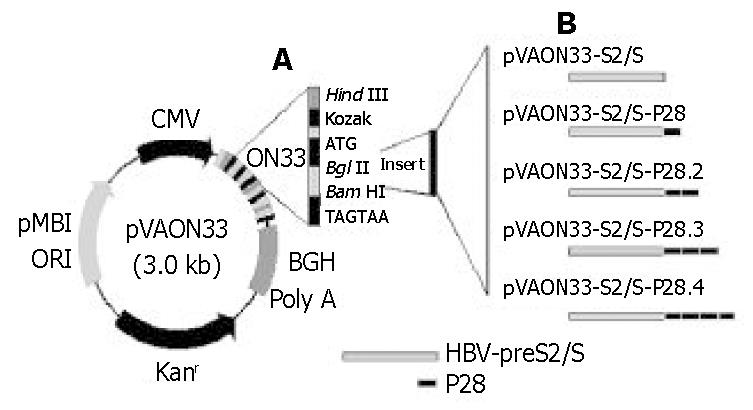
A eukaryotic expression vector pVAON33 was reconstructed from pVAX1, a kind gift from professor Zhongming Li (Food and Drug Administration, Bethesda, MD, USA). It contained the cytomegalovirus immediate-early (CMV-IE) promoter for initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation signal [BGH poly(A)] for termination of transcription. It also contained pMB1 origin of replication for prokaryotic replication as well as the kanamycin resistance gene (Kanr) for selection in antibiotic media. Moreover, a synthetic oligonucleotide (ON33) containing the Kozak’s translation initiation sequence was inserted into pVAX1 for the convenience of cloning four tandem repeats of the C3d-P28 in-frame with the gene encoding HBV-preS2/S (Figure 1). Plasmid pTG825 (tPA-C3d3) containing the encoding gene of C3d was a gift from Dr. Ted M. Ross (University of Pittsburgh, Pittsburgh, PA, USA). Plasmid pcDNA-preS2/S containing the encoding gene of HBV-preS2/S was constructed and conserved by our laboratory. pGEM-T vector was purchased from Huashun Biotechnology (Shanghai, China).
Human hepatocellular carcinoma cell lines SMMC-7721 was obtained from the Department of Biochemistry and Moleculr Biology, Shanghai Medical College, Fudan University (Shanghai, China). SMMC-7721 cells were grown in RPMI 1640 medium (Gibco BRL) supplemented with 100 mL/L calf bovine serum (CBS), 100 × 103 U/L penicillin and 100 × 103 U/L streptomycin sulfate. Eschrichia coli strain, DH5α was kept in our laboratory and maintained in LB medium.
Female BALB/c mice (H2d, 6 to 8-week-old) were purchased from the Center of Experimental Animal, Fudan University (Shanghai, China). They were randomly divided into 6 groups (6 mice per group) and fed with a standard laboratory diet under a specific pathogen-free (SPF) condition.
AMV reverse trancriptase, dNTP, Taq polymerase and PCR production-purified kits were purchased from Shanghai Biostar Co.. Hind III, BamH I and BglII restriction endonuclease, T4 DNA ligase, DNA marker 2000 and RNase-free DNase I were purchased from MBI Co. (Ukraine). Horseradish peroxidase-labeled goat anti-mouse IgG were purchased from Southern Biotech (USA). Lipofectamine was purchased from the Invitrogen Co. (USA). The anion-exchange resin columns were purchased from Qiagen (Germany). The synthetic oligonucleotide (ON) and the PCR primers used were as follows: ON33, 5’-AGCTTGCCACCATGAGATCTGGATCCTAGTAAG-3’ (sense) and 5’-GATCTTACTAGGATCCAGATCTCATGGTGGCA-3’ (antisense), which was used for reconstruction of the pVAX1 palsmid; C3d-P28, 5’-GAAGATCTAAGTTTCTGAACACAGC-3’ (sense) and 5’-CGGGATCCGGCCTAGGATGTG-3’ (antisense), which permitted the amplification of a 84-bp cDNA of C3d-P28; HBV-preS2/S, 5’-GGAAGATCTCAGTGGAATTCCACAAC-3’ (sense, bases 3082 to 3100) and 5’-GGAAGATCTAATGTATACCCAAAGACAA-3’ (antisense, bases 706 to 688), which permitted the amplification of the entire coding region of HBV-preS2/s cDNA; T7 primers, 5’-CGATGAAGATCTCT-3’ (sense) and 5’-CGATGAAGATCTCT-3’ (antisense), which were used for identification of recombinant plasmids by PCR; GAPDH primers, 5’-CTGCACCACCAACTGCTTAG-3’ (sense) and 5’-CCACTGACACGTTGGCAGTG-3’ (antisense), which permitted the amplification of a 275-bp band corresponding to human GAPDH transcripts as described previously.
The HBV-preS2/S gene was amplified by PCR from plasmid pcDNA-preS2/S. The gene encoding C3d-P28 amplified from plasmid pTG825 by PCR was cloned into the cloning vector-pGEM-T. Each gene was inserted into pVAON33 genetic immunization vector using standard molecular biological technology (Figure 1). Briefly, a 858 bp PCR-amplified preS2/S fragment was digested with BglII and ligated into the BglII restriction enzyme sites in the pVAON33. The vector expressing preS2/S-P28-fusion proteins was constructed by cloning four tandem repeats of the P28-encoding gene in-frame with preS2/S fragment and was designed based upon Dempsey et al[2]. Potential proteolytic cleavage sites between the junctions of HBV-preS2/S and P28 and the junction of P28 were mutated by ligating BamH I and BglII restriction endonuclease sites to replace an arginine codon with a glycine codon. All recombinant plasmids amplified in Eschrichia coli, DH5α, were verified by PCR, restricted endonucleases digestion and DNA sequencing, and then were purified using anion-exchange resin columns. Purity of DNA preparations was determined by optical density reading at 260 nm and 280 nm. All the plasmid DNA were stored at -20 °C in sterile saline at 1 mg/mL for experimental use.
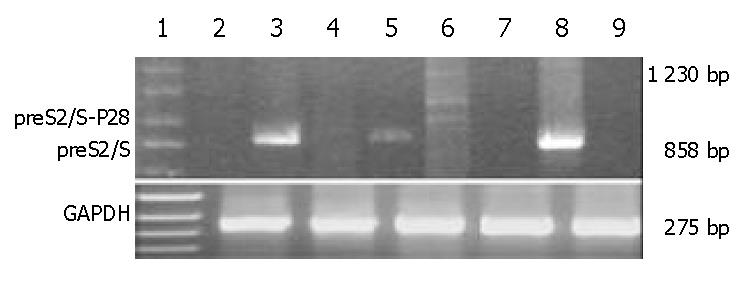
SMMC-7721 cells (5 × 105 cells), a human hepatocellular carcinoma cell line, were transfected with 2 μg of the different plasmids DNA mixed with 100 g/L lipofectamine according to the manufacturer’s guidelines. Semiquantitative RT-PCR was used to analyze the transcription of HBV-preS2/S or preS2/S-P28 fusions and ensure the correct size of fusions containing variable copies of C3d-P28. Total RNA was extracted from the transfected cells (1 × 106 cells) using guanidinium isothiocyanate-based buffer system according to the manufacturer’s instructions and then treated by RNase-free DNase I. First-strand cDNA was synthesized from 5 µL of total cellular RNA with a GeneAmp PCR system 2400 cycler (Perkin Elmer company, USA) using the following protocol: one cycle at 42 °C for 60 min and 70 for 10 min[16]. The cDNA was subjected to 30 cycles of amplification using the appropriate primers: (1) sense and antisense of HBV-preS2/S, which permitted the amplification of the HBV-preS2/S cDNA; (2) HBV-preS2/S sense and C3d-P28 antisense, which permitted the amplification of the preS2/S-P28 fusions cDNA; (3) sense and antisense of GAPDH, which permitted the amplification of the GAPDH cDNA. After initiate denaturation at 94 °C for 5 min, the amplification conditions were as follows: denaturation at 94 °C for 1 min, primers annealing at 58 °C for 1 min, and extension at 72 °C for 2 min. Final PCR products were analyzed on 10 g/L agarose gel electrophoresis and the intensity of the preS2/S and GAPDH PCR products was assessed by densitometry and quantitated by using UV-2000 (Tanon Science and Technology Ltd., China); preS2/S mRNA expression was calculated as the ratio of the intensity of the preS2/S band to the GAPDH band. At the same time, direct PCR reaction was performed to exclude the contamination of potential plasmid DNA in the RNA extracted from the transfected cells.
The quadriceps of mice were first injected with a total of 100 μL of 2.5 g/L bupivacaine to enhance the cellular uptake of plasmid DNA one day before the first immunization. Then 100 μg of each plasmid DNA dissolved in 100 μL of normal saline was injected into the same region. Twelve weeks later, each mouse was boosted by subcutaneous injection with 1.0 μg of purified HBsAg protein diluted in 100 μL of sterile PBS. Sera were collected at 2-week intervals after primary immunization for antibody analysis.
The anti-HBs antibody response elicited in mice was evaluated by ELISA. In brief, microtiter plates were coated with purified HBsAg protein (0.5 μg/well). After incubation with 100 mL/L FBS in PBS for 60 min at 37 °C to prevent nonspecific binding, 1:10 dilution of mouse serum were added to the plates and incubated at 37 °C for an additional 60 min. After the samples were washed with PBS containing 0.5 g/L Tween-20, a horseradish peroxidase-labeled goat anti-mouse antibody was added at a dilution 1:5000. After 60 min incubation, plates were washed, and substrate was added for color development. Absorbance (A490) of each plate wells was read at 490 nm that indicated the levels of specific antibody.
Relative antibody avidity was determined by a modified elution ELISA that was similar to serum antibody determination ELISAs up to the addition of samples as previously described[11,12]. Plates were washed 3 times with 0.5 g/L PBS-Tween 20. Sodium thiocyanate (NaSCN), a chaotropic compound that interferes with the antigen-antibody reaction, was added subsequently in concentrations ranging from 0.5 mol/L to 3.5 mol/L. Plates were allowed to left for 15 min at room temperature and then washed six times with PBS-Tween 20. Subsequent steps were performed similarly to the serum antibody determination ELISA. The avidity index (AI) corresponded to the effective concentration of NaSCN required to give a 50% reduction in absorbance at 490 nm.
Data were expressed as mean ± SD. Experimental results were analyzed by variance analysis or t test with SPSS software. P < 0.05 were considered statistically significant.
To test for an effect of C3d-P28 on the immune responses to a DNA vaccine, we selected plasmids encoding HBV envelope proteins as a model system. Then the genes encoding middle (preS2 plus S) HBV envelope proteins (858 bp) were cloned into a eukaryotic expression vector pVAON33. One to four tandem repeats of the C3d-P28 fragment were inserted into pVAON33 plasmid to construct the recombinant plasmids pVAON33-P28, pVAON33-P28.2, pVAON33-P28.3 and pVAON33-P28.4 (pVAON33-P28.[1-4]), respectively. HBV-preS2/S coding sequence was then introduced into the pVAON33-P28.[1-4], respectively. The results of identification by both PCR and DNA sequencing showed that the coding-codon of HBV-preS2/S coding gene, repeats of the P28-coding gene and their junctions were correct (data not shown).
SMMC-7721 cells were transiently transfected with preS2/S and preS2/S-P28.4 protein expression vectors, with the plasmid pVAON33 serving as a negative control. At 2 d after transfection, the preS2/S mRNA could be detected in SMMC-7721 cells transfected with both plasmid pVAON33-S2/S and pVAON33-S2/S-P28.4, but not in mock plasmid (Figure 2). However, mRNA representing fusions of preS2/S to various copies of P28 could only be detected in SMMC-7721 cells transfected with pVAON33-S2/S-P28.4 plasmid. Digested with DNase I, RNA samples extracted from the cells transfected with both pVAON33-S2/S and pVAON33-S2/S-P28.4 were confirmed the exclusion of potential plasmid DNA contamination (Figure 2). These results suggested that various copies of P28 had fused with the HBV-preS2/S. Semiquantitative RT-PCR analysis showed that preS2/S expressed by pVAON33-S2/S-P28.4-DNA was 29.61% lower than that expressed by pVAON33-S2/S-DNA.
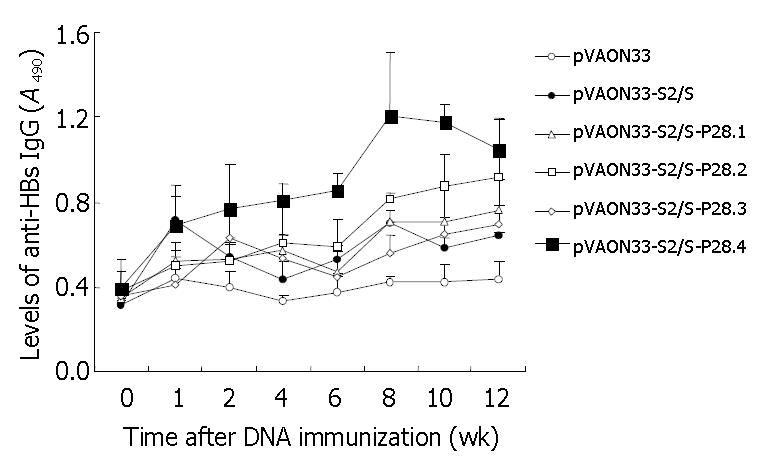
The preS2/S-P28-expressing DNA plasmids showed higher levels of anti-HBs antibody than the preS2/S-expressing DNA (Figure 3). When sera were assayed on HBsAg-coated plates, there were various degrees of enhancement of anti-HBs IgG antibody levels in the mice with DNA expressing fusions of HBV-preS2/S to variable copies of P28 compared to the mice primed with DNA expressing HBV-preS2/S only (Figure 3). Interestingly, at 12 wk after the primary immunization, the highest level of the specific antibody response (A490, 1.049 ± 0.186) was observed in the groups primed with DNA expressing fusions of HBV-preS2/S to four copies of P28, which was not only significant higher than the level of antibody response from the group primed with preS2/S-expressing DNA (A490, 0.621 ± 0.064; P < 0.01), but also obviously higher than the level of antibody response from the groups primed with preS2/S-P28.1-expressing DNA (A490, 0.764 ± 0.143; P < 0.01), preS2/S-P28.2-expressing DNA (A490, 0.915 ± 0.275; P < 0.05) and preS2/S-P28.3-expressing DNA (A490, 0.696 ± 0.091; P < 0.01), respectively. It was suggested that P28, when fused with HBV-preS2/S, could enhance the specific antibody response against HBsAg following primary DNA immunization. Differences in the enhancement of the antibody levels raised by the fusions of preS2/S-P28 to variable repeats of P28 appeared to be determined by the different repeats of P28, in which four copies of P28 might be necessary to achieve the most modest antibody response.
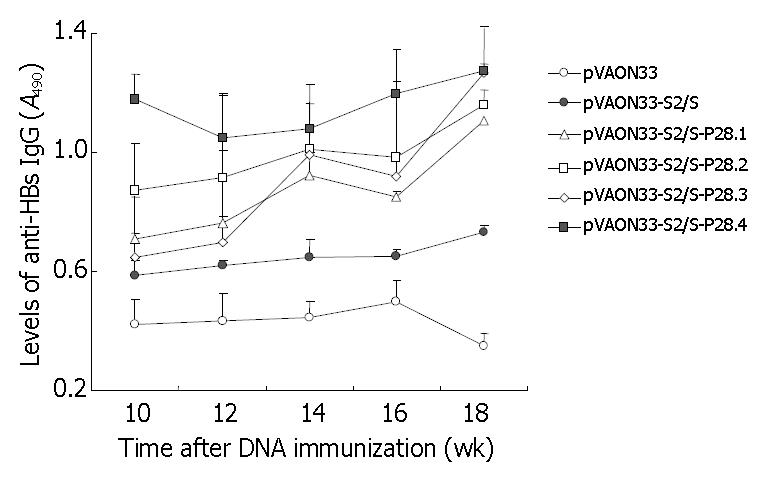
To further observe the effects of P28 peptides on the memory responses against HBsAg, mice from each group were boosted subcutaneously with 1 μg of HBsAg purified at the twelfth week after the primary immunization. The results showed that the mice of the groups vaccinated primarily with preS2/S-expressing DNA produced an increased levels of anti-HBs antibody compared to the mice immunized primarily with mock DNA after the second immunization with specific antigen (Figure 4). However, the acceleration of antibody levels was significantly higher and faster in the mice immunized with DNA expressing preS2/S-P28 fusions than that in the mice immunized with DNA expressing preS2/S only (P < 0.05). Interestingly, at the eighteenth week after the primary immunization, mice immunized primarily with DNA expressing preS2/S-P28.4 fusions maintained the highest levels of anti-HBs antibodies in all animals (A490, 1.273 ± 0.106). It was suggested that the mice immunized with DNA expressing preS2/S-P28 fusions not only enhanced primary humoral responses against HBsAg, but also produced a quicker and stronger memory responses than that with DNA expressing preS2/S only, in which four copies of P28 might also be necessary to obtain the most modest antibody memory reaction.
To observe whether P28 enhances the avidity maturation of the antibody response after immunization with P28-preS2/S fusions, NaSCN-displacement ELISA, a well-accepted approach, was used to evaluate the avidity of the anti-HBs antibodies[11-13]. The results demonstrated that the AI collected at 18 weeks from mice immunized primarily with preS2/S-P28.3 and preS2/S-P28.4 expressing DNA, separately was significantly higher than that from preS2/S-DNA vaccinated mice (Figure 5, P < 0.01). However, no significant increases in the AI were found in either preS2/S-P28.1 expressing or preS2/S-P28.2 expressing DNA vaccinated mice compared to the mice immunized with DNA expressing preS2/S only. These results suggested that C3d-P28 could enhance the avidity maturation of antibodies against HBsAg following gene immunization with DNA expressing fusions of HBV-preS2/S to three or four repeats of C3d-P28.
Recently, several studies demonstrated that DNA vaccines encoding the small or middle viral envelope proteins of HBV elicited higher titer antibody, as well as cell-mediated immune responses compared with HBV subunit vaccines[17-19]. Moreover, DNA immunization was able to break immune tolerance in the HBV transgenic mouse model[20] and induces protective antibody responses in human non-responders to conventional vaccination[17], indicating that DNA vaccines might be beneficial for immunotherapy for individuals with chronic HBV infections. DNA vaccines for HBV infection are efficacious, as well as inexpensive, and therefore, may be a promising tool in controlling the viral infections and liver diseases[21].
However, one of the drawbacks that prevents the wide spread use of DNA vaccines is slower rise in humoral immunity compared with other vaccine strategies for HBV infection. This may be due to the low levels of antigen expression and presentation by inoculated DNA plasmids compared to the inoculation of HBsAg[22]. In order to enhance the efficacy of antigens expressio n and presentation in the DNA immunization, several studies have shown that three copies of C3d, as a molecular adjuvant when fused to an antigen, could enhance the immune responses and accelerate the antibody avidity maturation[2-7]. However, Suradhat et al[23] reported that fusions of one or two copies of C3d with either bovine rotavirus VP7 or bovine herpesvirus type 1 glycoprotein D inhibited the specific humoral response following DNA immunization, and suggested that using three or more copies of C3d molecule may be necessary for efficient enhance of antigen-specific immune responses following DNA immunization. These results indicated that different repeats of C3d in the fusions of target antigen to C3d might influence efficient enhancement of antigen specific immune response following DNA immunization[2-7,23].
HBsAg is encoded by a single gene which is divided into S, preS2 and preS1 regions. preS vaccines have been most extensively applied in humans and shown to confer certain protective immunity. Recently more researches indicated that addition of the preS2 sequence could enhance the immunogenicity of the vaccines in human[24,25]. Therefore in this study, we selected HBV-preS2/S as a model antigen and different repeats of P28 peptide, an active peptide corresponding to the CR2-binding site on C3d[10-13], as a molecular adjuvant to explore enhancing effects on the immune response. As expected, mice vaccinated with DNA expressing fusions of preS2/S to P28 (one, two, three or four repeats) elicited a higher level of primary humoral response compared to mice vaccinated with DNA expressing only the preS2/S protein. Moreover, the mice immunized with DNA expressing preS2/S-P28 fusions produced a quicker and stronger memory responses than that with DNA expressing preS2/S only after second immunization. Interestingly, the results showed that the enhancing effect of various repeats of P28 in fusions of preS2/S to P28 on primary response as well as memory reaction was different, in which four copies of P28 might be necessary to achieve the most modest antibody response. It was indicated that the most modest antibody response appeared to be determined by the different repeats of P28. These results acquired here further explained the conflict between reports by Ross et al[4] and by Suradhat et al[23]. The increase in antibody response elicited by four copies of P28, similar to previous studies using antigen-C3d3 fusions[3-7], was even more intriguing, since the preS2/S-expression at the mRNA level by preS2/S-P28.4-expressing plasmid was 29.61% lower than the expression by plasmid expressing HBV-preS2/S only. The most likely mechanism by which P28 increases antibody responses is similar to C3d, which has been hypothesized to reflect the binding of C3d to CD21 on the surface of B-cells or follicular dendritic cells (FDC)[3,4,8].
Surprisingly, immunization with the preS2/S-P28 fusions also resulted in enhanced avidity maturation of anti-HBs antibody. The results demonstrated that the AI of sera collected from mice immunized primarily with preS2/S-P28.3 and preS2/S-P28.4 expressing DNA, separately, was significantly higher than that from preS2/S-DNA vaccinated mice. Avidity is a term used to describe the strength of interaction between multivalent antigens and antibodies in serum and is dependent on the individual affinities of the polyclonal antibodies[3]. Avidity maturation occurs in germinal centers where the somatic hypermutation of immunoglobulin results in a large repertoire of Ag-specific B cells that undergo inducing or selection for high-affinity B cell receptors by T helper cells or follicular dendritic cells (FDC)[8,9,26-28]. The most likely mechanism by which P28 enhanced the avidity maturation of anti-HBs antibodies might relate to multiple interactions of cells responsible for humoral immunity, including B cells, T helper cells, and FDC. These interactions, coupled with the enhancing effects of CR2 ligation on germinal center formation[29] and the enhancing effects of specific Th cells on somatic hypermutations of B cells[30], could promote the selection of antigen-specific B cells with high-affinity B cell receptors, resulting in the avidity maturation of anti-HBs antibodies and the development and maintenance of memory B cells[31].
Taken together, this is the first report to demonstrate that immunization with DNA expressing antigen-P28 fusions results in enhancing antibody response and enhancing antibody avidity maturation. These findings argue that four or more repeats of C3d-P28 may be necessary for efficient enhancement of antigen-specific immune responses, and thus C3d-P28, a short, active peptide derived from the CR2-binding site on complement C3d, may be another promising molecular adjuvant and may assist the future design of vaccines for health-threatening pathogens.
We sincerely thank Dr. T.M. Ross from University of Pittsburgh School of Medicine for helpful advice in this study and for providing the plasmid. We also thank Jin-Ping Zhang, Cong-Feng Xu, Huan-Bin Xu for their technical assistance.
Edited by Kumar M Proofread by Xu FM
| 1. | Pepys MB. Role of complement in induction of antibody production in vivo. Effect of cobra factor and other C3-reactive agents on thymus-dependent and thymus-independent antibody responses. J Exp Med. 1974;140:126-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 264] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 2. | Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 874] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 3. | Test ST, Mitsuyoshi J, Connolly CC, Lucas AH. Increased immunogenicity and induction of class switching by conjugation of complement C3d to pneumococcal serotype 14 capsular polysaccharide. Infect Immun. 2001;69:3031-3040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Ross TM, Xu Y, Bright RA, Robinson HL. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1:127-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 165] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Mitchell JA, Green TD, Bright RA, Ross TM. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine. 2003;21:902-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Ross TM, Xu Y, Green TD, Montefiori DC, Robinson HL. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with gp120-C3d fusion proteins. AIDS Res Hum Retroviruses. 2001;17:829-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Green TD, Newton BR, Rota PA, Xu Y, Robinson HL, Ross TM. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine. 2001;20:242-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Tew JG, Wu J, Qin D, Helm S, Burton GF, Szakal AK. Follicular dendritic cells and presentation of antigen and costimulatory signals to B cells. Immunol Rev. 1997;156:39-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 199] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 9. | Yellin MJ, Sinning J, Covey LR, Sherman W, Lee JJ, Glickman-Nir E, Sippel KC, Rogers J, Cleary AM, Parker M. T lymphocyte T cell-B cell-activating molecule/CD40-L molecules induce normal B cells or chronic lymphocytic leukemia B cells to express CD80 (B7/BB-1) and enhance their costimulatory activity. J Immunol. 1994;153:666-674. [PubMed] |
| 10. | Lambris JD, Ganu VS, Hirani S, Müller-Eberhard HJ. Mapping of the C3d receptor (CR2)-binding site and a neoantigenic site in the C3d domain of the third component of complement. Proc Natl Acad Sci USA. 1985;82:4235-4239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Esparza I, Becherer JD, Alsenz J, De la Hera A, Lao Z, Tsoukas CD, Lambris JD. Evidence for multiple sites of interaction in C3 for complement receptor type 2 (C3d/EBV receptor, CD21). Eur J Immunol. 1991;21:2829-2838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Servis C, Lambris JD. C3 synthetic peptides support growth of human CR2-positive lymphoblastoid B cells. J Immunol. 1989;142:2207-2212. [PubMed] |
| 13. | Frade R, Hermann J, Barel M. A 16 amino acid synthetic peptide derived from human C3d triggers proliferation and specific tyrosine phosphorylation of transformed CR2-positive human lymphocytes and of normal resting B lymphocytes. Biochem Biophys Res Commun. 1992;188:833-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Sarrias MR, Franchini S, Canziani G, Argyropoulos E, Moore WT, Sahu A, Lambris JD. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J Immunol. 2001;167:1490-1499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Lou D, Kohler H. Enhanced molecular mimicry of CEA using photoaffinity crosslinked C3d peptide. Nat Biotechnol. 1998;16:458-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Chow YH, Chiang BL, Lee YL, Chi WK, Lin WC, Chen YT, Tao MH. Development of Th1 and Th2 populations and the nature of immune responses to hepatitis B virus DNA vaccines can be modulated by codelivery of various cytokine genes. J Immunol. 1998;160:1320-1329. [PubMed] |
| 17. | Rottinghaus ST, Poland GA, Jacobson RM, Barr LJ, Roy MJ. Hepatitis B DNA vaccine induces protective antibody responses in human non-responders to conventional vaccination. Vaccine. 2003;21:4604-4608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Kim SJ, Suh D, Park SE, Park JS, Byun HM, Lee C, Lee SY, Kim I, Oh YK. Enhanced immunogenicity of DNA fusion vaccine encoding secreted hepatitis B surface antigen and chemokine RANTES. Virology. 2003;314:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Davis HL, Mancini M, Michel ML, Whalen RG. DNA-mediated immunization to hepatitis B surface antigen: longevity of primary response and effect of boost. Vaccine. 1996;14:910-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 119] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 20. | Mancini M, Hadchouel M, Davis HL, Whalen RG, Tiollais P, Michel ML. DNA-mediated immunization in a transgenic mouse model of the hepatitis B surface antigen chronic carrier state. Proc Natl Acad Sci USA. 1996;93:12496-12501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Hassett DE, Whitton JL. DNA immunization. Trends Microbiol. 1996;4:307-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 84] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Robinson HL, Pertmer TM. DNA vaccines for viral infections: basic studies and applications. Adv Virus Res. 2000;55:1-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 89] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Suradhat S, Braun RP, Lewis PJ, Babiuk LA, van Drunen Littel-van den Hurk S, Griebel PJ, Baca-Estrada ME. Fusion of C3d molecule with bovine rotavirus VP7 or bovine herpesvirus type 1 glycoprotein D inhibits immune responses following DNA immunization. Vet Immunol Immunopathol. 2001;83:79-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Davis HL, Brazolot Millan CL. DNA-based immunization against hepatitis B virus. Springer Semin Immunopathol. 1997;19:195-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 25. | Jilg W. Novel hepatitis B vaccines. Vaccine. 1998;16 Suppl:S65-S68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 671] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 27. | Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 828] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 28. | Fang Y, Xu C, Fu YX, Holers VM, Molina H. Expression of complement receptors 1 and 2 on follicular dendritic cells is necessary for the generation of a strong antigen-specific IgG response. J Immunol. 1998;160:5273-5279. [PubMed] |
| 29. | Griffioen AW, Rijkers GT, Janssens-Korpela P, Zegers BJ. Pneumococcal polysaccharides complexed with C3d bind to human B lymphocytes via complement receptor type 2. Infect Immun. 1991;59:1839-1845. [PubMed] |
| 30. | Razanajaona D, van Kooten C, Lebecque S, Bridon JM, Ho S, Smith S, Callard R, Banchereau J, Brière F. Somatic mutations in human Ig variable genes correlate with a partially functional CD40-ligand in the X-linked hyper-IgM syndrome. J Immunol. 1996;157:1492-1498. [PubMed] |
| 31. | Fischer MB, Goerg S, Shen L, Prodeus AP, Goodnow CC, Kelsoe G, Carroll MC. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science. 1998;280:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 202] [Article Influence: 7.5] [Reference Citation Analysis (0)] |