Published online Jun 1, 2004. doi: 10.3748/wjg.v10.i11.1600
Revised: December 4, 2003
Accepted: December 8, 2003
Published online: June 1, 2004
AIM: To characterize the gene expression profiles associated with activation of mouse hepatic stellate cell (HSC) and provide novel insights into the pathogenesis of hepatic fibrosis.
METHODS: Mice HSCs were isolated from BALB/c mice by in situ perfusion of collagenase and pronase and single-step density Nycodenz gradient. Total RNA and mRNA of quiescent HSC and culture-activated HSC were extracted, quantified and reversely transcripted into cDNA. cDNAs from activated HSC were labeled with Cy5 and cDNAs from the quiescent HSC were labeled with Cy3, which were mixed with equal quantity, then hybridized with cDNA chips containing 4000 genes. Chips were washed, scanned and analyzed. Increased expression of 4 genes and decreased expression of one gene in activated HSC were confirmed by reverse transcription- polymerase chain reaction (RT-PCR).
RESULTS: A total of 835 differentially expressed genes were identified by cDNA chip between activated and quiescent HSC, and 465 genes were highly expressed in activated HSC. The differentially expressed genes included those involved in protein synthesis, cell-cycle regulation, apoptosis, and DNA damage response.
CONCLUSION: Many genes implicated in intrahepatic inflammation, fibrosis and proliferation were up-regulated in activated HSC. cDNA microarray is an effective technique in screening for differentially expressed genes between two different situations of the HSC. Further analysis of the obtained genes will help understand the molecular mechanism of activation of HSC and hepatic fibrosis.
- Citation: Liu XJ, Yang L, Luo FM, Wu HB, Qu-Qiang. Association of differentially expressed genes with activation of mouse hepatic stellate cells by high-density cDNA mircoarray. World J Gastroenterol 2004; 10(11): 1600-1607
- URL: https://www.wjgnet.com/1007-9327/full/v10/i11/1600.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i11.1600
Liver fibrosis is a common consequence of chronic liver injury and is characterized by the progressive accumulation of extracellular matrix (ECM) proteins, particularly type I and III collagens. Hepatic stellate cells (HSC) are the major source of ECM in hepatic fibrosis and HSC is one of the sinusoid-constituent cells that plays multiple roles in the liver pathophysiology. After hepatic injury, HSC undergoes an activation process, characterized by loss of vitamin A, trans-differentiation to a smooth muscle a-actin (a-SMA) -positive myofibroblast like cell type, increased proliferation and increased production of ECM proteins[1,2]. Activation and transformation of HSC from the vitamin A-storing phenotype (also called “quiescent” phenotype) to the “myofibroblastic” one has been identified as a critical step in hepatic fibrogenesis and is regulated by several factors including cytokines and oxidative stress[3-5]. However, the molecular mechanism for HSC activation is not well understood. The activation of HSC involves many genes from multiple pathogenic pathways.
cDNA microarray analysis is a powerful descriptive method of examining the expression profile of hundreds to thousands of genes in unison. It has become an increasingly popular tool to investigate the function of genes, especially those genes involved in tumor generation and growth[6]. Recently, cDNA array has been used to identify differentially expressed genes in HCV-associated cirrhosis and achieve new insights into HCV liver injury[7].
Further advances in our knowledge about HSC activation requires more genes to be identified. Microarray technology provides us with a genomic approach to explore the genetic markers and molecular mechanisms leading to hepatic fibrosis. To this end, we have used cDNA microarray analysis to detect genes whose mRNA expression changes in the cultured activated HSC. RT-PCR analysis confirmed up-regulation of 4 previously unreported transcripts and down-regulation of one gene transcript in the activated HSC. The identification of these genes provides new insight into the understanding of HSC activation and hepatic fibrogenesis. Culturing HSC on plastic surface converts them from a quiescent phenotype to an activated phenotype similar to in vivo activation and this cultured-induced activation has been extensively studied as a model of the activation secondary to liver fibrogenesis[8,9]. Therefore, we used the in vitro model in which the activation of HSC was induced by growth on plastic dishes to study the differentially expressed genes associated with the activation of HSC.
Male BALB/c mice were obtained from Experimental Animal Center of West China Medical School, Sichuan University (Chengdu, Sichuan). All animals were treated humanely according to the national guideline for the care of animals.
Pronase, DNase I and Collagenase B were from Roche Molecular Biochemicals (Mannhein, Germany). Nycodenz was from Sigma (St. Louis, USA). Dulbecco’s modified medium (DMEM), trypsin-EDTA and new-born calf serum were from Invitrogen Corp (Grand Island, USA). Monoclonal antibodies against desmin, a-smooth muscle actin (a-SMA) were obtained from Dako (Glostrup, Denmark). Gene chips (MGEC-40s) were purchased from BioStar Genechip Inc. (Shanghai, P.R.China), and each chip contains 4000 mouse cDNAs, including 1500 cDNAs of known sequence and function, and 2500 novel cDNAs whose function has not been known in the public database.
HSC isolation and culture HSC was isolated from male Balb/c mice by in situ pronase, collagenase perfusion and single-step Nycodenz gradient according to our previous report. The purity of primary HSC after 3 d in culture was approximately 95% as estimated by vitamin A auto-fluorescence and immunocytochemistry with antibody against desmin. Therefore, HSC cultured in uncoated plastic dishes spontaneously acquired an activated phenotype, characterized by expression of α-SMA and by loss of vitamin A droplets. After reaching confluence (about 10-14 d after plating), activated HSC was detached by trypsin, and split in a 1:2 ratio. Experiments were performed on primary cells cultured for 3 d and activated HSC of the third passages using 3 independent cell lines, and the purity of activated HSC exceeded 98%.
Preparation of RNA and cDNA microarray Total RNA was isolated from primary mouse HSC and sub-confluent culture-activated mouse HSC (passage 3), using Trizol reagent (Invitrogen Life Technologies Inc, USA) according to the manufacturer’s protocol. Poly (A) mRNA was isolated from total RNA using Oligotex mRNA Midi Kit (Qiagen, USA) according to the manufacturer’s protocol.
All microarray procedures were performed by BioStar Genechip Inc. (Shanghai, P.R.China). Equal quantities of mRNA from each cell phenotype were used to prepare probes, hybridized to gene chips (MGEC-40 s, Biostar Genechip Inc.), and analyzed for the quantity of mRNA encoded by 4000 mouse genes. The preparation of Cy5 and Cy3 probes from mRNA samples and the hybridization were conducted by the BioStar Genechip Inc.
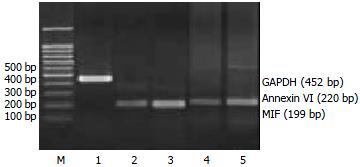
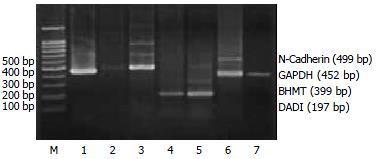
RT-PCR assays To validate the expression pattern identified on the expression arrays, 4 genes (MIF, Annexin VI, N-Cadherin, DAD1) from the up-regulated genes and one gene (BHMT) from the down-regulated genes in activated HSC were picked and semi-quantitative RT-PCR was performed to confirm their changed expression with cDNA templates from activated and quiescent HSC. The total RNA was isolated from HSC using Trizol reagent, precipitated in ethanol and resuspended in sterile RNAase-free water and stored at -70 °C, as described previously. One-step reverse transcription-polymerase chain reaction (RT-PCR) was performed according to the method of the supplier (TaKaRa Biotechnology Co., Ltd, Dalian). Primers were designed using the Primer 3 program from Whitehead Institute for Biomedical Research (Cambridge, MA, USA)[10], synthesized and purified by PAGE in Genebase BRL Custom Primers (Genebase Biotechnology Co., Shanghai). Primer sequences are shown in Table 1. The PCR products was analyzed by 20 g/L agarose gel electrophoresis with TAE buffer at 80 V for 40 min, visualized with ethidium bromide and photographed under UV light by Gel Documentation system (Gel Doc 2000TM, Bio-Rad, USA). The semi-quantitative analysis was performed using the volume analysis in the Quantity One Software (Bio-Rad, USA). Each detected gene/GAPDH quotient is the indication of the detected gene. Experiments were performed for at least five times with similar results.
| Gene name | Primer sequence | Expected product size |
| Betaine homocysteine | Left 5’–GCC TAT AGC GGC TAC CAT GT-3’ | 399 bp |
| methyltransferase (BHMT) | Right 5’-CTC TGC AAT GGC CCT GAT GT-3’ | |
| Defender against cell | Left 5’-TTC GGC TAC TGT CTC CTC GT -3’ | 197 bp |
| death protein 1 (DAD1) | Right 5’-ACG AGG TGC AGG ATA GTG CT-3’ | |
| Annexin VI | Left 5’-TAC CCC GGA GTA TTT TGC TG-3’ | 220 bp |
| Right 5’-GTC CCC TCC ACA TAG CTT CA-3’ | ||
| Neural cadherin | Left 5’-ATA CAG TGT CAC TGG GCC AG- 3’ | 499 bp |
| (N-Cadherin) | Right 5’-CGT AAG TGG GAT TGC CTT CC- 3’ | |
| Macrophage migration | Left 5’-TTT TAG TGG CAC GAG CGA CC-3’ | 199 bp |
| inhibitory factor (MIF) | Right 5’-AAG CGA AGG TGG AAC CGT TC- 3’ | |
| GAPDH | Left 5’-ACC ACA GTC CAT GCC ATC AC-3’ | 452 bp |
| Right 5’-TCC ACC ACC CTG TTG CTG TA-3’ |
RT-PCR results were expressed as mean ± SD. Differences between means were analyzed with Student t test for paired samples. A value of P < 0.05 was considered statistically significant.
Differences in gene expression patterns between mouse activated HSC and quiescent HSC were assessed using microarray analysis. This array allows a quantitative measurement of 4000 known genes and expressed sequence tags. Genes that differed in intensity by at least 2-fold were considered to be differentially regulated. Figure 1 shows that the cDNA array images along with color charts indicating up-regulated genes with red, down-regulated ones with green and non-changed with yellow. Of the 4000 genes analyzed by mircoarray, a total of 835 genes (20.8%) revealed differential expression in the activated HSC when compared with the quiescent HSC (Table 2, Table 3, Table 4). Of the 835 genes with altered expression in the activated HSC, 462 genes (including 204 known function genes) revealed elevated expression whereas 373 genes (including 132 known function genes) revealed reduced expression. Array analysis identified many differentially expressed genes that are important in inflammation, fibrosis, proliferation, signaling, apoptosis and oxidative stress.
| GenBank accession number | Gene name | Ratio (Cy5/Cy3) | Potential gene function |
| M73741 | Alpha-B2-crystallin, complete cds | 155.619 | Small heat shock protein gene, an early marker for HSC activation |
| X52046 | Procollagen, type III, alpha 1 | 119.092 | Extracellular matrix (ECM), over- expressed in hepatic fibrosis |
| M18194 | Fibronectin mRNA | 29.958 | ECM, over expressed in hepatic fibrosis |
| J02870 | Laminin receptor mRNA | 6.62 | ECM receptor (integrin), cell adhesion |
| L08115 | CD9 antigen | 6.094 | Cell membrane glycoprotein, involved in cell activation and adhesion |
| U16163 | Prolyl 4-hydroxylase alpha (II) - subunit mRNA, complete cds | 4.555 | An enzyme which is essential for the collagen synthesis in HSC |
| X62622 | TIMP-2 mRNA for tissue inhibitor of metalloproteinase, type 2 | 3.732 | Inhibition for ECM degradation |
| X04017 | mRNA for cysteine-rich glycoprotein SPARC | 3.435 | Regulation of cell shape, adhesion, migration and proliferation |
| AF070470 | SPARC-related protein (SRG) mRNA, complete cds | 3.181 | Regulation of cell shape, adhesion, migration |
| M21495 | Cytoskeletal gamma-actin mRNA, | 3.092 | Relate skeleton structure of cell |
| AF188297 | TGF-beta receptor binding protein (Trip1) mRNA, complete cds | 2.976 | TGF-beta mediated signaling pathway |
| AJ245923 | alpha-tubulin 8 (Tuba 8 gene) mRNA | 2.901 | Relate skeleton structure of cell |
| AF053454 | Tetraspan TM4SF (Tspan-6) mRNA complete cds, | 2.896 | Cell development, differentiation, motility |
| AF013262 | Lumican gene, complete cds | 2.862 | Small, leucine-rich proteoglycan which involves in the regulation of collagen figril assembly |
| L02526 | Protein kinase (MEK) mRNA | 2.714 | Signaling molecule in MAPK family |
| Y00769 | mRNA for integrin beta subunit | 2.521 | Signaling molecular in cell adhesion, proliferation and migration |
| GenBank accession number | Gene name | Ratio (Cy5/Cy3) | Potential gene function |
| AF061017 | UDP-glucose dehydrogenase, mRNA complete cds | 15.56 | Glycosaminoglycan , hyaluronan and heparin sulfate biosynthesis |
| X65553 | mRNA for poly (A) binding protein | 7.405 | A regulator of translation initiation |
| AF029844 | Elongation factor 1-beta homology mRNA, complete cds | 6.305 | Translation factor and has multi-functions |
| NM_010798 | Macrophage migration inhibitory factor (Mif), mRNA | 5.847 | Proinflammatory peptide and a mediator of growth factor dependent ERK MAP kinase activation and cell cycle progression |
| M31131 | Neural cadherin (N-cadherin) mRNA | 5.559 | Cell adhesion molecule |
| AB029930 | mRNA for caveolin-1 beta isoform | 4.945 | Inhibit the eNOS activity |
| D12618 | mRNA for nucleosome assembly protein-1, complete cds | 4.528 | Histone H2A-H2B shutting protein that promotes histone deposition, and is important for maintaining chromatin structure |
| M22432 | Protein synthesis elongation factor Tu (eEF-Yu, eEf-1-1alpha) mRNA, complete cds | 4.344 | Translation factor involved in the protein biosynthesis |
| D31717 | mRNA for ribophorin | 4.319 | Glycoprotein synthesis |
| NM_012052 | Ribosomal protein S3 (Rps3), mRNA | 4.297 | Protein biosynthesis |
| X72711 | mRNA for replication factor C (140 Kda), long subunit | 4.245 | DNA synthesis and repair, and a regulator of NF-kappa B |
| X13460 | Annexin VI, p68 (Mouse mRNA for p68 protein of the lipocortin family) | 4.218 | Calcium binding protein |
| NM_011290 | Ribosomal protein L6 (Rp16), mRNA | 4.089 | Protein biosynthesis |
| U12273 | Apurinic/apyrimidinic endonuclease (APEX) gene, complete cds | 3.488 | A DNA repair enzyme and an activator of several transcription factors |
| D42048 | mRNA for squalene epoxidase | 3.298 | Rate-limiting enzyme of cholesterol biosynthesis |
| AJ250491 | mRNA for receptor activity modifying protein 3 (Ramp3) gene | 3.255 | Complex with the calcitonin receptor-like receptor, establishing a functional receptor for adrenomedullin |
| L04280 | Ribosomal protein (RPL12) mRNA | 3.172 | Protein biosynthesis |
| K02927 | Ribonucleotide reductase subunit M1 mRNA, complete cds | 3.156 | Rate-limiting enzyme in DNA synthesis and repair |
| AF087568 | Palmitoyl-protein thioesterase precursor mRNA, complete cds | 3.059 | Cell survival signaling |
| AF020039 | NADP-dependent isocitrate dehydrogenase (Idh) mRNA, complete cds | 3.038 | A key enzyme of the tricarboxylic acid cycle |
| X57960 | Ribosomal protein L7 | 2.962 | Protein biosynthesis |
| U09816 | GM2 activator protein (Gm2a) mRNA | 2.888 | Enzymatic hydrolysis of GM2 |
| AB018575 | Cdc 7 mRNA, complete cds | 2.816 | Essential for G1/S transition |
| AF041054 | E1B19K/Bcl-2 binding protein homology (Nip3) mRNA, nuclear gene encoding mitochondria protein, complete cds | 2.813 | Nips is a pro-apoptotic mitochondria protein |
| L04128 | Ribosomal protein L18 (rpL18) mRNA | 2.751 | Protein biosynthesis |
| Z31554 | (129/SV) Cctd mRNA for CCT (chaperonin containing TCP-1) delta subunit | 2.743 | Stabilize the cytoskeletal protein actin and tubulin |
| D63784 | mRNA for MIDA1, complete cds | 2.703 | Regulate cell growth |
| AF026481 | eIF-1A (eIF-1A) mRNA, complete cds | 2.682 | Translation initiation factor |
| U35249 | CDK-activating kinase assembly factor p36/MAT1, complete cds | 2.645 | An assembly factor and a targeting subunit of cyclin-dependent kinase-activating kinase that is involved in cell cycle control, transcription and DNA repair |
| M76131 | Elongation factor (ef-2) mRNA, 3’end | 2.614 | Protein biosynthesis |
| X64713 | mRNA for cyclin B1 | 2.576 | Cell cycle regulatory protein |
| D26091 | mRNA for mCDC47, complete cds | 2.570 | A member of the minichromosome maintenance (Mcm) family involve in the DNA replication licensing system |
| U78085 | Ribosomal protein S5 mRNA | 2.507 | Protein synthesis |
| AF141322 | Caveolin-2 mRNA, complete cds | 2.458 | Inhibit the eNOS activity |
| M33934 | IMP dehydrogenase mRNA, complete cds | 2.429 | An enzyme involved in de novo synthesis of guanine nucleotides |
| U83628 | Defender against cell death protein 1 (DAD1) mRNA, complete cds | 2.372 | Apoptotic suppressor gene |
| GenBank accession number | Gene name | Ratio (Cy5/Cy3) | Potential gene function |
| D49949 | mRNA for IGIF precursor poly-peptide in (Interleukin 18), complete cds | 0.065 | In the development of Th1 cells and tissue injury inflammatory reaction |
| U63146 | Retinal binding protein (RBP) mRNA | 0.125 | Retinoid storage and metabolism |
| AF035644 | Potentially prenylated protein tyrosine phosphatase mPRL-2 mRNA, complete cds | 0.226 | Cellular regulation |
| X58287 | mR-PTPu gene for protein tyrosine phosphatase, receptor-type M | 0.280 | Cellular regulation |
| AF033381 | Betaine homocysteine methyl transferase (BHMT) mRNA, complete cds | 0.299 | A key liver enzyme that is important for homocysteine homeostasis |
| M75720 | Alpha-1 serine protease inhibitor 3 mRNA, complete cds | 0.308 | Anti-inflammatory effect |
| U20497 | P19 protein mRNA, complete cds | 0.324 | CDK4&CDK6 inhibitor, cell cycle inhibitor |
| D85596 | AMP deaminase H-type, complete cds | 0.339 | Involved in the biosynthesis, inter-conversion and degradation of purine compounds |
| Y10138 | Gene encoding prostaglandin D synthase | 0.361 | A PGD producing enzyme and a retinoid transporter |
| NM_013498 | CAMP response element modulator (CREM) mRNA | 0.366 | Down-regulator of CAMP-induced transcription |
| U67187 | G protein signaling regulator RGS2 (rgs2) mRNA, complete cds | 0.369 | Negatively regulate G-coupled receptor function |
| AF023919 | PK-120 precursor (itih-4) mRNA, complete cds | 0.421 | Inter-alpha-trypsin-inhibitor H4, is a potential regulator for ECM proteins |
| AF077950 | Protein inhibitor of activated STAT protein PIAS 1 mRNA, complete cds | 0.433 | Inhibition of STAT-1-mediated gene activation |
| M75718 | Alpha-1 protease inhibitor 4 mRNA, complete cds | 0.437 | Anti-inflammatory activity |
| D38046 | mRNA for type II DNA topoisomerase beta isoform, complete cds | 0.445 | An essential enzyme that alters DNA topology which is important for cell survive and apoptosis |
| AF073996 | myotubularin (Mtm1) mRNA complete cds | 0.464 | Subfamily of protein tyrosine phosphatases |
To further investigate the reliability of our array data, we picked 5 differential expressed genes and measured the expression of the genes in the activated and quiescent HSC. Figures 2 and 3 and Table 5 show that the different expression pattern of each of the five genes as determined by RT-PCR were similar to those observed with cDNA array, confirming the reliability of our array data.
| Groups | Mean ± SD | N | p value |
| (quotient of the detected gene/GAPDH) | |||
| MIF from quiescent HSC | 0.269 ± 0.016 | 5 | |
| MIF from activated HSC | 0.759 ± 0.046 | 5 | < 0.05 |
| N-Cadherin from quiescent HSC | 0.177 ± 0.063 | 5 | |
| N-Cadherin from activated HSC | 0.776 ± 0.087 | 5 | < 0.05 |
| Annexin VI from quiescent HSC | 0.137 ± 0.04 | 5 | |
| Annexin VI from activated HSC | 0.478 ± 0.025 | 5 | < 0.05 |
| DAD1 from quiescent HSC | 0.174 ± 0.016 | 5 | |
| DAD1 from activated HSC | 0.593 ± 0.047 | 5 | < 0.05 |
| BHMT from quiescent HSC | 0.602 ± 0.083 | 5 | |
| BHMT from activated HSC | 0.134 ± 0.059 | 5 | < 0.05 |
Genome-wide expression profiling by microarray of cDNA or oligonucleotide probes on a glass or nylon substrate is an exceptionally powerful tool for the study of gene regulation. This methodology has been used to investigate the phenomena particularly appropriate for the analysis of expressed liver genes[7]. cDNA microarrays were used to profile changes in gene expression in activated HSC. Scatter plot analysis showed that approximately 20.8% of all mouse genes examined on the 4000 gene microarrays exhibited altered expression, with 11.5% showing up-regulation and 9.3% showing down-regulation. These genes will be future studied.
In the up-regulated genes associated with the activation of HSC, some genes have already been reported (Table 2). Alpha B-crystallin was first reported by Lang et al[11] recently as an early marker for HSC activation. In our experiment, the ratio of Cy5 to Cy3 for its mRNA was the highest in all the genes in the gene-chip, suggesting that mRNA expression of alpha B-crystallin in activated HSC up-regulated mostly comparing with the quiescent HSC. The mRNA expression for procollagen type III[12], fibronectin[13], laminin receptor, prolyl 4-hydroxylase[14], TIMP-2, TGF-beta and its receptor binding protein[15], MEK[16] and integrin beta[17] was increased in activated HSC. Tetraspanins (TM4SF) super family which includes CD9, CD53, CD81 and CD151 were highly expressed in the activated human HSC and have been implicated in HSC migration, a key event in liver tissue wound healing and fibrogenesis[18]. Secreted protein, acidic and rich in cysteine (SPARC), which functions in tissue remodeling, was expressed by activated HSC in chronic hepatitis, suggesting the involvement of SPARC in hepatic fibrogenesis after chronic injuries[19]. Lumican is a small leucine-rich proteoglycan, which contributes to cell migration, proliferation, tissue hydration and collagen fibrillogenesis, and its expression is increased in HSC in diseased liver during the process of fibogenesis[20]. Although we employed different methods, animal models or different sources of HSC, our experimental data was consistent with the results observed by others, demonstrating that the technique of cDNA microarray has a higher reliability. In addition, the expression of those that were not previously linked to the activation of HSC was also found to be changed. These include genes involved in the control of HSC morphology, growth, differentiation, migration and apoptosis.
Analysis of the genes showed elevated expression in the activated HSC clustered into distinct functional groups. Genes showing elevated expression included many genes involved in the formation and remodeling of the extracellular matrix (ECM) and in the regulation of cellular response (including cell adhesion, proliferation and migration) to the ECM, such as procollagen type III, fibronectin, TIMP-2, TGF beta, UDP-glucose dehydrogenase (AF061017), N-cadherin (M131131) and lumican (AF013262).
The enzyme UDP-glucose dehydrogenase (Udpgdh) (EC 1.1.1.22) converts UDP-glucose to UDP-glucutonate, a critical component of the glycosaminoglycans, hyluronan, chondroitin sulfate and heparan sulfate[21]. It is known that heparan sulfate proteoglycans are essential cofactors in cell-matrix adhesion processes, in cell-cell recognition system, and in receptor-growth factor interactions. Cultured human HSC can synthesize all four cyndecans and the increased expression of glycosaminoglycans and hyaluronic acid may be important in the deposition of matrix components and activation of growth factors accompanying fibrogenesis. Our results suggested that the increased expression of heparan sulfate proteoglycans in activated HSC might be partially caused by the elevated expression of the enzyme UDP-glucose dehydrogenase.
Neural cadherin (N-cadherin) is an adhesion molecule of the cadherin family, whose expression was up regulated in response of smooth muscle cells to arterial injury[22]. AnnexinVI is a 68-Kda protein of the annexin family, a group of structural similar, calcium-dependent, phospholipid-binding proteins[23]. Our cDNA microarray data along with the RT-PCR results showed increased expression of N-cadherin and annexin VI in activated HSC, suggesting their regulations might be important for the hepatic fibrogenesis.
In all cells, protein synthesis is coordinated by the ribosome, and the large ribonucleoprotein is composed of at least 50 distinct molecules and several large RNA molecules. Genes showing elevated expression included many genes that encode ribosomal proteins and proteins involved in the translation and protein synthesis, such as mRNA for poly A-binding protein (X65553), elongation factor 1-beta homology mRNA (AF029844), elongation factor Tu (M22432), ribosomal protein S3 (NM_012052), S5 (U78085), L6 (NM_011290), L12 (L04280), L18 (L04128) and eukaryotic initiation factor 1A (eIF1A) (AF026481). HSC proliferated during the process of activation, so it is not surprising that many genes involved in the protein synthesis up-regulated in the activated HSC.
Our work has identified some previously unreported genes involved in DNA synthesis and repair showing elevated expression in the activated HSC, such as replication factor C gene (X72711), apurinic/apyrimidinic endonuclease (APEX) gene (U12273), ribonucleotide reductase subunit M1 (M1-RR) mRNA (K02917), and IMP dehydrogenase mRNA (M33934), an enzyme involved in de novo synthesis of guanine nucleotides.
Replication factor C (RFC) is a clamp loader, catalyzes assembly of circular proliferating cell nuclear antigen clamps around primed DNA, enabling processive synthesis by DNA polymerase during DNA replication and repair. The Rel A (p65) subunit of NF-kappa B is an important regulator of inflammation, proliferation and apoptosis, but the large subunit of RFC can function as a regulator of Rel A. In addition to its previously described function in DNA replication and repair, RFC plays an important role as a regulator of transcription factor NF-kappa B activity[24].
APEX nuclease is a mammalian DNA repair enzyme having apurinic/apyrimidinic endonuclease, 3’-5’-exonuclease, DNA 3’ repair diesterase and DNA 3’-phosphatase activities. It is also a redox factor (Ref-1), stimulating DNA binding activity of AP-1 binding proteins such as Fos and Jun[25]. Ribonucleotide reductase (RR) is a cytoplasmatic enzyme catalyzing the reduction of all four ribonucleotides to their corresponding deoxyribonucleotides, so it is a rate-limiting enzyme in the DNA synthesis and repair. Its activity strongly correlates to the rate of DNA synthesis, and the expression of M1-RR antigen was found to correlate positively with the expression of Ki-67 and PCNA, the cell cycle markers of proliferating cells[26]. We conclude that mechanisms for DNA synthesis and repair are activated during the process of HSC activation.
Elevated expression was also observed for a number of genes involved in the control of cell growth, survival, differentiation and apoptosis. Palmitoyl-protein thioesterase (PPT) is a newly described lysomal enzyme that hydrolyzes long chain fatty acids from lipid-modified cysteine residues in proteins, and its precursor mRNA (AF087568) was up-regulated in activated HSC. It was reported that inhibition of PPT increased the susceptibility of neurons to apoptotic cell death[27]. Id, a helix-loop-helix protein not only regulates cell differentiation negatively, but also promote growth and apoptosis, and an Id-associate protein, MIDA1 (Mouse Id associate1), regulated cell growth positively. MIDA1 is a novel sequence-specific DNA binding protein with some different properties from the usual transcription factors and may act as a mediator of Id-mediated growth-promoting function through its DNA binding activity[28], and its gene expression was increased in the activated HSC.
The cyclin-dependent kinase (CDK) -activating kinase (CAK) is involved in cell cycle control, transcription, and DNA repair, and MAT1 gene (U35249), an assembly factor and a targeting subunit of CAK, was also up regulated in the activated HSC. It was reported that abrogation of MAT1 expression by retrovirus-mediated gene transfer of antisense MAT1 RNA in cultured rat aortic smooth muscle cells (SMC) retarded SMC proliferation and inhibits cell activation from a nonproliferation state, and this effect was due to G1 phase arrest and apoptotic cell death[29].
Up-regulation of the DAD1 (U83628), a putative anti-apoptosis gene identified in several distantly related organisms[28], was observed as well as the Nip3 (AF041054), a proapoptotic member of the Bcl-2 family of cell death factors[29] in the activated HSC. CDC7, an evolutionarily conserved serine-threonine kinase, plays a pivotal role in linking cell cycle regulation to genome duplication, being essential for the firing of DNA replication origins[30]. Our microarray experiments also identified elevated expression for CDC7 gene (AB018575) and cyclin B1 (X64713) in the activated HSC. The strategy for terminating the proliferation of activated HSC by apoptosis might be an exciting therapy for patients with chronic liver injury and fibrosis[10,31], therefore, our experimental data about the differentially expressed genes involved in apoptosis will give some new ideas on induce apoptosis in HSC.
Macrophage migration inhibitory factor (MIF) (NM_010798), a pro-inflammatory peptide and a mediator of growth factor-dependent ERK MAP kinase activation and cell cycle progression[32], was up-regulated by the process of HSC activation. MIF has been shown to contribute significantly to the development of immuno-pathology in several models of inflammatory, such as glomerulonephritis[33]. Our RT-PCR results confirmed the increased expression of MIF mRNA in the activated HSC. This is the first study to demonstrate that activated HSC can produce MIF in vitro, and its up-regulation in activated HSC might suggest a role for MIF in the hepatic fibrogenesis in vivo. It is necessary to carry further more research to understand how MIF regulates proliferation in activated HSC.
Down-regulation was observed for genes encoding interleukin 18 (D49949) and retinal binding protein (RBP) (U63146) in the activated HSC. IL-18 has an anti-fibrotic effect and it was reported that intrasplenic transplantation of IL-18 gene modified hepatocytes could be a candidate for therapeutic intervention in hepatic fibrosis through induction of a dominant Th1 response[34]. HSCs are the body’s major cellular storage sites for retinoid, but the immortalized rat HSC cell line HSC-T6 failed to express RBP[35]. Our cDNA microarray results were consistent with the previous reports.
In the activated HSC, reduced expression was observed for genes involved in general cellular regulation including a family of protein-tyrosine phosphatase (PTPases) and some negative regulators of cell growth signaling, such as P19 protein (U20497), CAMP response element modulator (Crem) (NM_013498), G protein signaling regulator RGS2 (U67187), and protein inhibitor of activated STAT protein - PIAS1 (AF077950). The PTPases included the potentially prenylated protein tyrosine phosphatase mPRL-2 (AF035644)[36], myotubularin (Mtm1) (AF071996)[37] and mR-PTPu gene for protein tyrosine phosphatase, receptor type M. Protein tyrosine kinase and phosphatase play diverse roles in involving energy metabolism, cell proliferation and stimulation of MHC class I molecule pathway. Down-regulation was also observed for the alpha-1 serine protease inhibitor 3 (M75720), alpha-1 protease inhibitor 4 (M75718) and PK-120 precursor (itih-4) (AF023919), a serine protease inhibitor.
P19 is a tumor suppressing protein and belongs to a family of cyclin D-dependent kinase inhibitors of CDK4 and CDK6, which play a key role in human cell cycle control[38]. Addition of p19 protein can lead to inhibition of the CDK’s activity and may cause the cells to arrest at G1 phase. Transcriptional factors binding to camp-response elements (CREs) in the promoters of various genes belong to the basic domain-leucine zipper super family and are composed of three genes in mammals, CREB, CREM, and ATF-1. Activation is classically brought about by signaling-dependent phosphorylation of a key acceptor site (Ser133 in CREB) by a number of possible kinases, including PKA, CamKIV and RSK-2. Repression may involve dynamic dephosphorylation of the activators and decreased association with CREB-binding protein (CBP). Another pathway of transcriptional repression on CRE sites implicates the inducible repressor ICER (inducible camp early repressor), a product of the CREM gene. Being an inducible repressor, ICER is involved in auto-regulatory feedback loops of transcription that govern the down-regulation of early response genes, such as the proto-oncogene c-fos[39]. It is known that CREB is one of the transcription factors whose expression is increased in the activated HSC during the liver injury[40], but the important role of CREM in the pathophysiology of liver fibrogenesis has not been studied. Similarly, Jak-Stat signaling is one of the signaling pathways in the HSC proliferation and activation[41], but the role of a negative regulator in this cytokine signaling, protein inhibitor of activated STAT-1 (PIAS-1), has not been understood. Thus, our microarray data might provide novel potential approaches to the treatment of hepatic fibrogenesis in patients with chronic liver diseases.
Reduced mRNA expression was also found for betaine-homocysteine methyl transferase (BHMT). BHMT is a key liver enzyme for homocysteine (Hcy) homeostasis. It catalyzes the synthesis of methionine from betaine and homocysteine, utilizing a zinc ion to activate Hcy. Elevated plasma levels of Hcy have been shown to interfere with normal cell function in a variety of tissues and organs, such as the vascular wall and the liver. It is known that Hcy is able to induce the expression and synthesis of the TIMP-1 in variety of cell types ranging from vascular smooth muscle cells to hepatocytes, HepG2 cells and HSCs. In HSCs, Hcy also stimulates alpha1 (I) procollagen mRNA expression, promotes activating protein-1 (AP-1) binding activity[42]. Hcy is a key metabolite in methionine metabolism, which takes place mainly in the liver. Hyperhomocysteinemia may develop as a consequence of defects in Hcy-metabolizing genes (such as BHMT) ; nutritional conditions leading to vitamin B (6), B (12), or folate deficiencies; or chronic alcohol consumption. We postulated that hyperhomocysteinemia in the hepatic fibrosis was partly due to the reduced expression of BHMT gene in the activated HSC.
We used cDNA array analysis to detect genes whose mRNA expression changes in the activated mouse HSC after culture on the plastic dishes. RT-PCR analysis confirmed the up-regulation of four previously unreported transcripts and down-regulation of one gene in activated HSC. The identity of these genes provides new insights into the understanding of activation of HSC during the liver injury and hepatic fibrogenesis.
Edited by Ma JY Proofread by Xu FM
| 1. | Reeves HL, Friedman SL. Activation of hepatic stellate cells--a key issue in liver fibrosis. Front Biosci. 2002;7:d808-d826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 353] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 2. | Dai WJ, Jiang HC. Advances in gene therapy of liver cirrhosis: a review. World J Gastroenterol. 2001;7:1-8. [PubMed] [Cited in This Article: ] |
| 3. | Mann DA, Smart DE. Transcriptional regulation of hepatic stellate cell activation. Gut. 2002;50:891-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 158] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 4. | Pinzani M, Marra F. Cytokine receptors and signaling in hepatic stellate cells. Semin Liver Dis. 2001;21:397-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 326] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 5. | Liu XJ, Yang L, Mao YQ, Wang Q, Huang MH, Wang YP, Wu HB. Effects of the tyrosine protein kinase inhibitor genistein on the proliferation, activation of cultured rat hepatic stellate cells. World J Gastroenterol. 2002;8:739-745. [PubMed] [Cited in This Article: ] |
| 6. | Tan ZJ, Hu XG, Cao GS, Tang Y. Analysis of gene expression profile of pancreatic carcinoma using cDNA microarray. World J Gastroenterol. 2003;9:818-823. [PubMed] [Cited in This Article: ] |
| 7. | Shackel NA, McGuinness PH, Abbott CA, Gorrell MD, McCaughan GW. Insights into the pathobiology of hepatitis C virus-associated cirrhosis: analysis of intrahepatic differential gene expression. Am J Pathol. 2002;160:641-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 8. | Liu C, Gaça MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721-11728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Galli A, Crabb DW, Ceni E, Salzano R, Mello T, Svegliati-Baroni G, Ridolfi F, Trozzi L, Surrenti C, Casini A. Antidiabetic thiazolidinediones inhibit collagen synthesis and hepatic stellate cell activation in vivo and in vitro. Gastroenterology. 2002;122:1924-1940. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 334] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 10. | Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365-386. [PubMed] [Cited in This Article: ] |
| 11. | Lang A, Schrum LW, Schoonhoven R, Tuvia S, Solís-Herruzo JA, Tsukamoto H, Brenner DA, Rippe RA. Expression of small heat shock protein alphaB-crystallin is induced after hepatic stellate cell activation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1333-G1342. [PubMed] [Cited in This Article: ] |
| 12. | Wei HS, Li DG, Lu HM, Zhan YT, Wang ZR, Huang X, Zhang J, Cheng JL, Xu QF. Effects of AT1 receptor antagonist, losartan, on rat hepatic fibrosis induced by CCl (4). World J Gastroenterol. 2000;6:540-545. [PubMed] [Cited in This Article: ] |
| 13. | Svegliati-Baroni G, Ridolfi F, Di Sario A, Saccomanno S, Bendia E, Benedetti A, Greenwel P. Intracellular signaling pathways involved in acetaldehyde-induced collagen and fibronectin gene expression in human hepatic stellate cells. Hepatology. 2001;33:1130-1140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 113] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Aoyagi M, Sakaida I, Suzuki C, Segawa M, Fukumoto Y, Okita K. Prolyl 4-hydroxylase inhibitor is more effective for the inhibition of proliferation than for inhibition of collagen synthesis of rat hepatic stellate cells. Hepatol Res. 2002;23:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Tahashi Y, Matsuzaki K, Date M, Yoshida K, Furukawa F, Sugano Y, Matsushita M, Himeno Y, Inagaki Y, Inoue K. Differential regulation of TGF-beta signal in hepatic stellate cells between acute and chronic rat liver injury. Hepatology. 2002;35:49-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 158] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | Kim KY, Rhim T, Choi I, Kim SS. N-acetylcysteine induces cell cycle arrest in hepatic stellate cells through its reducing activity. J Biol Chem. 2001;276:40591-40598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 112] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Yang C, Zeisberg M, Mosterman B, Sudhakar A, Yerramalla U, Holthaus K, Xu L, Eng F, Afdhal N, Kalluri R. Liver fibrosis: insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology. 2003;124:147-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 212] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 18. | Mazzocca A, Carloni V, Sciammetta S, Cordella C, Pantaleo P, Caldini A, Gentilini P, Pinzani M. Expression of transmembrane 4 superfamily (TM4SF) proteins and their role in hepatic stellate cell motility and wound healing migration. J Hepatol. 2002;37:322-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | Nakatani K, Seki S, Kawada N, Kitada T, Yamada T, Sakaguchi H, Kadoya H, Ikeda K, Kaneda K. Expression of SPARC by activated hepatic stellate cells and its correlation with the stages of fibrogenesis in human chronic hepatitis. Virchows Arch. 2002;441:466-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Gressner AM, Krull N, Bachem MG. Regulation of proteoglycan expression in fibrotic liver and cultured fat-storing cells. Pathol Res Pract. 1994;190:864-882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Ge X, Penney LC, van de Rijn I, Tanner ME. Active site residues and mechanism of UDP-glucose dehydrogenase. Eur J Biochem. 2004;271:14-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Jones M, Sabatini PJ, Lee FS, Bendeck MP, Langille BL. N-cadherin upregulation and function in response of smooth muscle cells to arterial injury. Arterioscler Thromb Vasc Biol. 2002;22:1972-1977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | de Diego I, Schwartz F, Siegfried H, Dauterstedt P, Heeren J, Beisiegel U, Enrich C, Grewal T. Cholesterol modulates the membrane binding and intracellular distribution of annexin 6. J Biol Chem. 2002;277:32187-32194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 24. | Anderson LA, Perkins ND. Regulation of RelA (p65) function by the large subunit of replication factor C. Mol Cell Biol. 2003;23:721-732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Ranalli TA, Tom S, Bambara RA. AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J Biol Chem. 2002;277:41715-41724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 58] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 26. | Chen S, Zhou B, He F, Yen Y. Inhibition of human cancer cell growth by inducible expression of human ribonucleotide reductase antisense cDNA. Antisense Nucleic Acid Drug Dev. 2000;10:111-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Dawson G, Dawson SA, Marinzi C, Dawson PE. Anti-tumor promoting effects of palmitoyl: protein thioesterase inhibitors against a human neurotumor cell line. Cancer Lett. 2002;187:163-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Inoue T, Shoji W, Obinata M. MIDA1 is a sequence specific DNA binding protein with novel DNA binding properties. Genes Cells. 2000;5:699-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Wu L, Chen P, Shum CH, Chen C, Barsky LW, Weinberg KI, Jong A, Triche TJ. MAT1-modulated CAK activity regulates cell cycle G (1) exit. Mol Cell Biol. 2001;21:260-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 30. | Kim JM, Nakao K, Nakamura K, Saito I, Katsuki M, Arai K, Masai H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002;21:2168-2179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Abriss B, Hollweg G, Gressner AM, Weiskirchen R. Adenoviral-mediated transfer of p53 or retinoblastoma protein blocks cell proliferation and induces apoptosis in culture-activated hepatic stellate cells. J Hepatol. 2003;38:169-178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Liao H, Bucala R, Mitchell RA. Adhesion-dependent signaling by macrophage migration inhibitory factor (MIF). J Biol Chem. 2003;278:76-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Fingerle-Rowson G, Koch P, Bikoff R, Lin X, Metz CN, Dhabhar FS, Meinhardt A, Bucala R. Regulation of macrophage migration inhibitory factor expression by glucocorticoids in vivo. Am J Pathol. 2003;162:47-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 99] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 34. | Zhang LH, Pan JP, Yao HP, Sun WJ, Xia DJ, Wang QQ, He L, Wang J, Cao X. Intrasplenic transplantation of IL-18 gene-modified hepatocytes: an effective approach to reverse hepatic fibrosis in schistosomiasis through induction of dominant Th1 response. Gene Ther. 2001;8:1333-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, Blaner WS. An immortalized rat liver stellate cell line (HSC-T6) : a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882-893. [PubMed] [Cited in This Article: ] |
| 36. | Si X, Zeng Q, Ng CH, Hong W, Pallen CJ. Interaction of farnesylated PRL-2, a protein-tyrosine phosphatase, with the beta-subunit of geranylgeranyltransferase II. J Biol Chem. 2001;276:32875-32882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 359] [Cited by in F6Publishing: 382] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 38. | Zeeb M, Rösner H, Zeslawski W, Canet D, Holak TA, Balbach J. Protein folding and stability of human CDK inhibitor p19 (INK4d). J Mol Biol. 2002;315:447-457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res. 2002;275:143-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 139] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Eng FJ, Friedman SL. Transcriptional regulation in hepatic stellate cells. Semin Liver Dis. 2001;21:385-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Saxena NK, Ikeda K, Rockey DC, Friedman SL, Anania FA. Leptin in hepatic fibrosis: evidence for increased collagen production in stellate cells and lean littermates of ob/ob mice. Hepatology. 2002;35:762-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 290] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 42. | García-Tevijano ER, Berasain C, Rodríguez JA, Corrales FJ, Arias R, Martín-Duce A, Caballería J, Mato JM, Avila MA. Hyperhomocysteinemia in liver cirrhosis: mechanisms and role in vascular and hepatic fibrosis. Hypertension. 2001;38:1217-1221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 88] [Article Influence: 3.8] [Reference Citation Analysis (0)] |