Copyright
©The Author(s) 2025.
World J Gastroenterol. Jun 28, 2025; 31(24): 108021
Published online Jun 28, 2025. doi: 10.3748/wjg.v31.i24.108021
Published online Jun 28, 2025. doi: 10.3748/wjg.v31.i24.108021
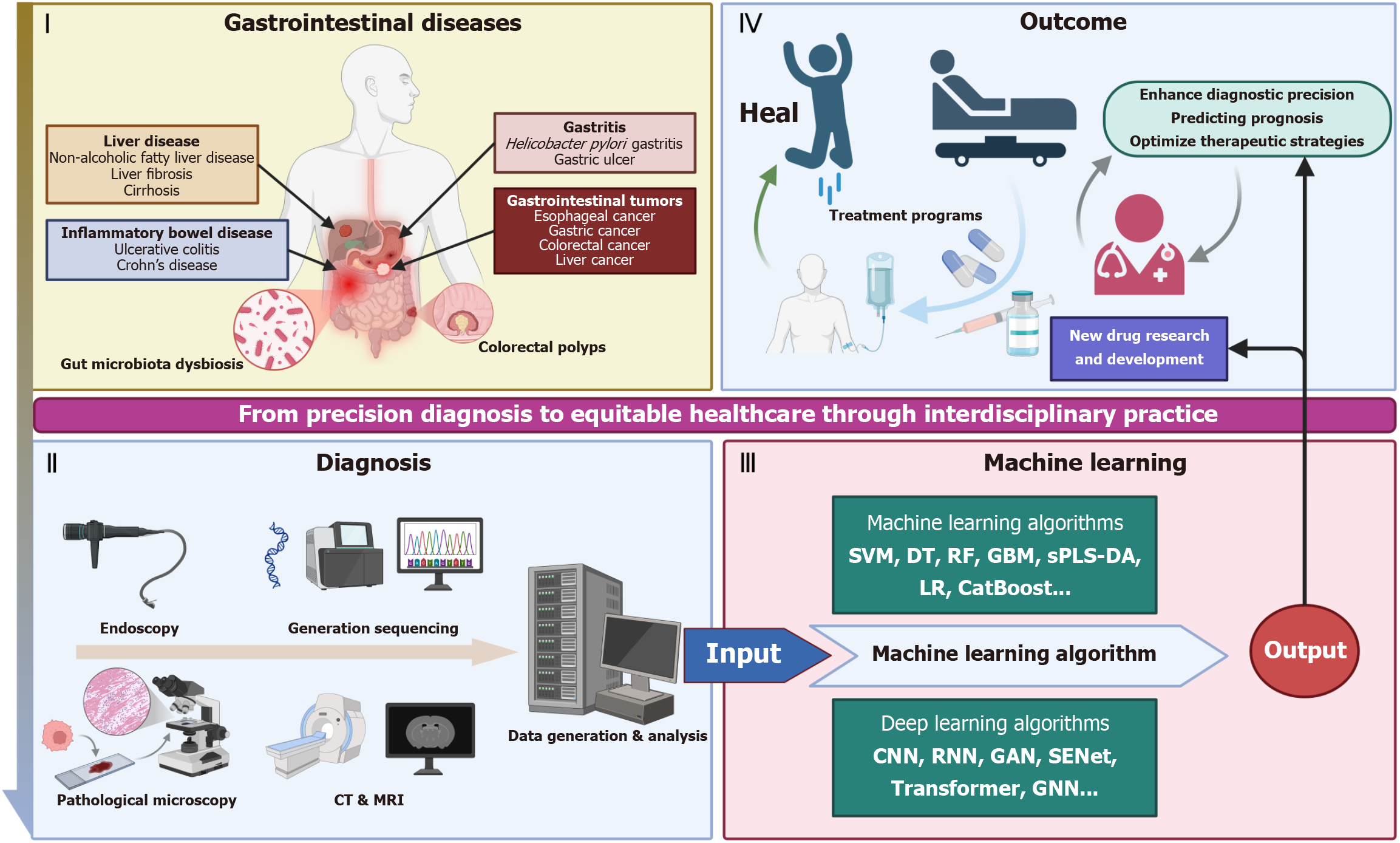
Figure 1 Artificial intelligence in gastrointestinal diseases.
Section I (gastrointestinal diseases) includes liver diseases (metabolic dysfunction-associated steatotic liver disease, liver fibrosis, cirrhosis), inflammatory bowel disease (ulcerative colitis, Crohn’s disease), gastritis (Helicobacter pylori gastritis, erosive mucosal changes), gastrointestinal tumors (esophageal cancer, gastric cancer, colorectal cancer, liver cancer), and colorectal polyps, with gut microbiota dysbiosis as a contributing factor. Section II (diagnosis) involves techniques such as endoscopy, generation sequencing, pathological microscopy, computed tomography and magnetic resonance imaging, followed by data generation and analysis. Section III (machine learning) presents algorithms: Machine learning algorithms (support vector machines, decision tree, random forest, gradient boosting machine, sparse partial least squares-discriminant analysis, logistic regression, CatBoost) and deep learning algorithms (convolutional neural network, recurrent neural network, generative adversarial network, squeeze-and-excitation network, transformer, graph neural network), which process input data. Section IV (outcome) demonstrates the therapeutic cycle: Treatment programs lead to healing or hospital care, promoting new drug research and development. Ultimately, it enhances diagnostic precision, predicts prognosis, and optimizes therapeutic strategies, embodying the journey from precision diagnosis to equitable healthcare via interdisciplinary practice. CT: Computed tomography; MRI: Magnetic resonance imaging; SVM: Support vector machines; DT: Decision tree; RF: Random forest; GBM: Gradient boosting machine; sPLS-DA: Sparse partial least squares-discriminant analysis; LR: Logistic regression; CNN: Convolutional neural network; RNN: Recurrent neural network; GAN: Generative adversarial network; SENet: Squeeze-and-excitation network; GNN: Graph neural network.
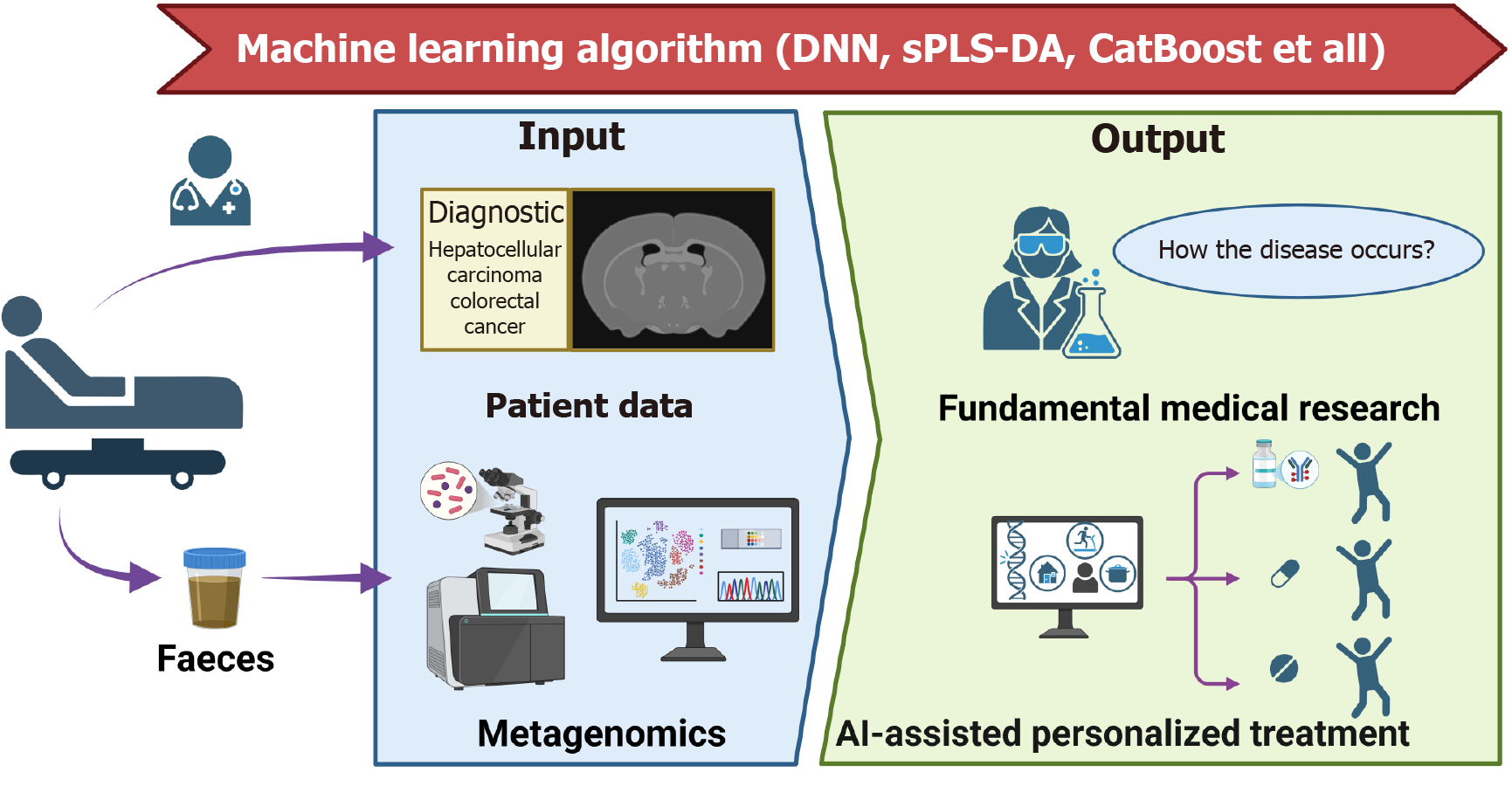
Figure 2 Schematic illustration of the deep integration between gut microbiome and machine learning for decoding cross-disease features to drive precision treatment.
In the “Input” section, patient data (e.g., diagnostic information on hepatocellular carcinoma and colorectal cancer) and metagenomics data (faeces samples) are utilized. Various machine learning algorithms, such as deep neural network, sparse partial least squares-discriminant analysis, and CatBoost-process these inputs. The “Output” involves two key dimensions: One is fundamental medical research exploring “How the Disease Occurs?” through gut microbiome pattern analysis; the other is artificial intelligence-assisted personalized treatment, which leverages the integration of microbiomics and artificial intelligence to formulate tailored therapeutic strategies. This synergy between metagenomics and artificial intelligence significantly promotes in-depth disease mechanism research and personalized patient treatment. sPLS-DA: Sparse partial least squares-discriminant analysis; AI: Artificial intelligence; DNN: Deep neural network.
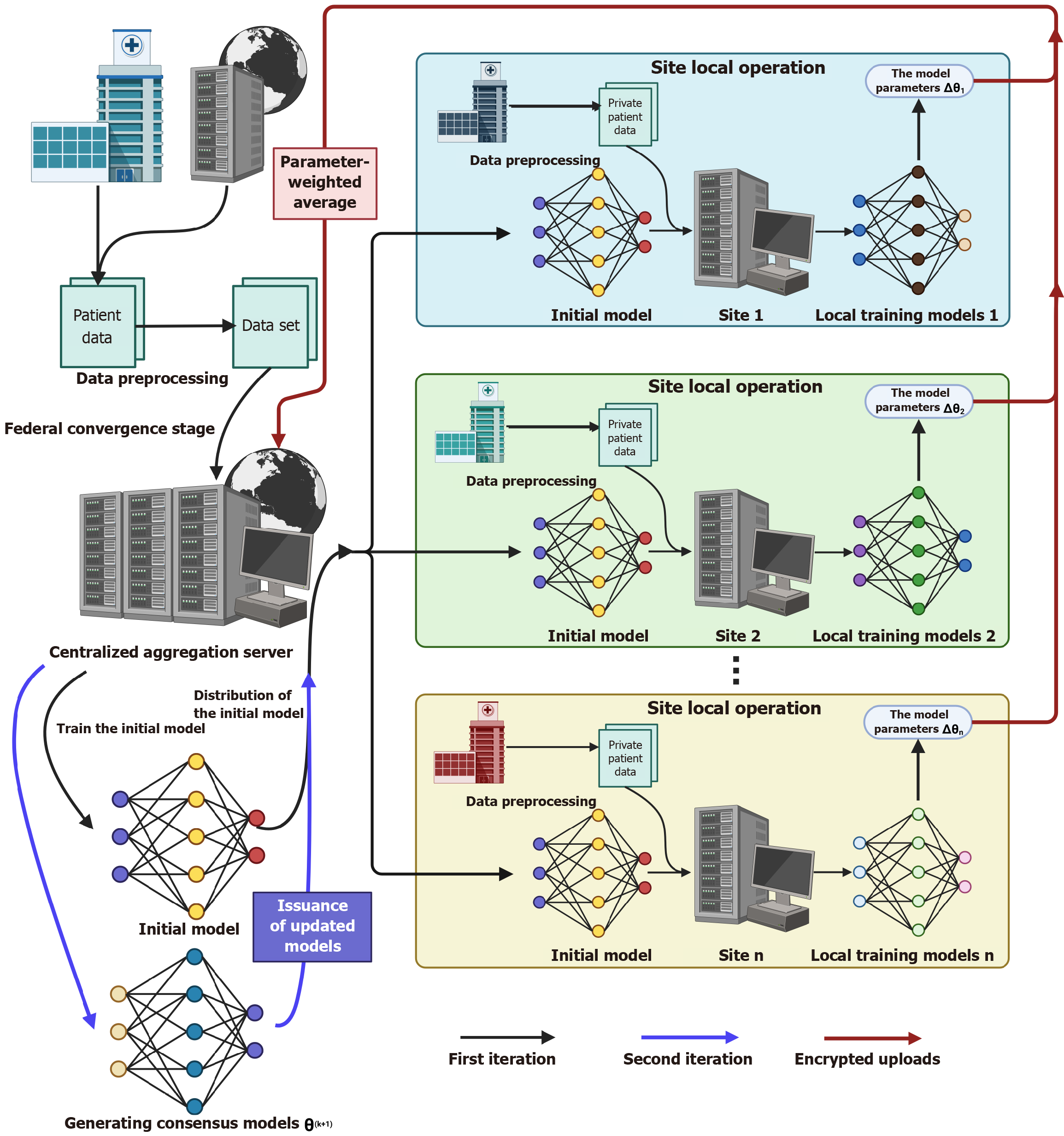
Figure 3 Schematic diagram of the envisioned federated learning architecture for gastrointestinal applications.
In the federated convergence stage, the centralized aggregation server first generates an initial model, which is distributed to multiple independent sites (e.g., site 1, site 2, and site n). Each site performs data preprocessing on private gastrointestinal patient datasets and conducts local training using the initial model to produce local training models. To emphasize data privacy protection, model parameters (marked as Δθi) are encrypted during upload to the centralized aggregation server. The server adopts parameter-weighted average to aggregate these parameters, issue updated models, and promote iterative training (illustrated by the first and second iterations). This architecture not only protects the privacy of gastrointestinal patient data through decentralized data processing and encryption strategies but also enhances model generalization. By integrating heterogeneous gastrointestinal datasets from multiple sites for collaborative training, the final consensus model θ (k + 1) can better adapt to diverse clinical scenarios. As a key framework, federated learning enables privacy-preserving collaborative model optimization, holding substantial promise for advancing artificial intelligence applications in gastrointestinal research.
- Citation: Chen ZL, Wang C, Wang F. Revolutionizing gastroenterology and hepatology with artificial intelligence: From precision diagnosis to equitable healthcare through interdisciplinary practice. World J Gastroenterol 2025; 31(24): 108021
- URL: https://www.wjgnet.com/1007-9327/full/v31/i24/108021.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i24.108021