Copyright
©The Author(s) 2016.
World J Gastroenterol. Feb 28, 2016; 22(8): 2475-2482
Published online Feb 28, 2016. doi: 10.3748/wjg.v22.i8.2475
Published online Feb 28, 2016. doi: 10.3748/wjg.v22.i8.2475
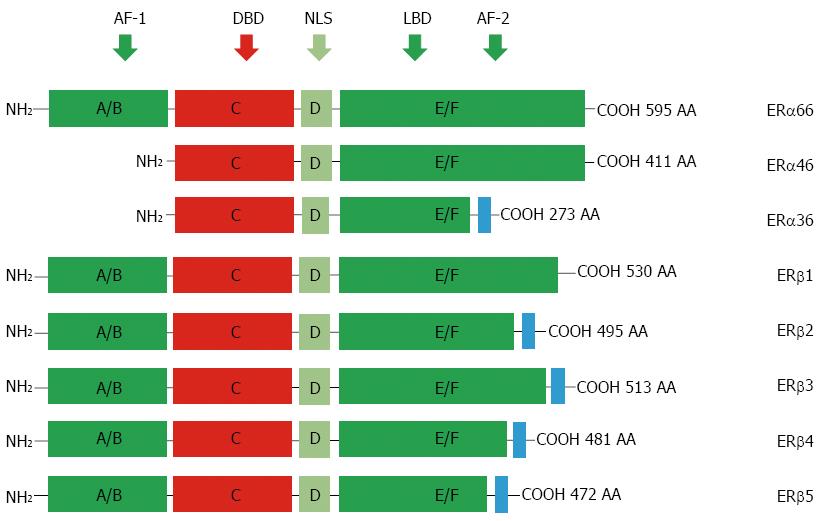
Figure 1 Protein structure of estrogen receptors.
Two different forms of ER are encoded by two distinct genes located in chromosomes 6 and 14 and produce two proteins with 595 and 530 amino acids in full length, respectively. Six evolutionary conserved domains, namely A-F, are shared by different ERs. For ERα isoforms, compared to the full length ERα66, ERα46 lacks AF-1 domain (A/M), ERα36 lacks AF-1 and partial AF-2 domains but is equipped an extra different C-terminal. For alternatively-spliced ERβ isoforms, they differ mainly at their C-terminals. AF-1: Transcriptional activation factor-1; DBD: DNA-binding domain; NLS: Nuclear localization signals; LBD: Ligand binding domain; ERs: Estrogen receptors; AA: Amino acid.
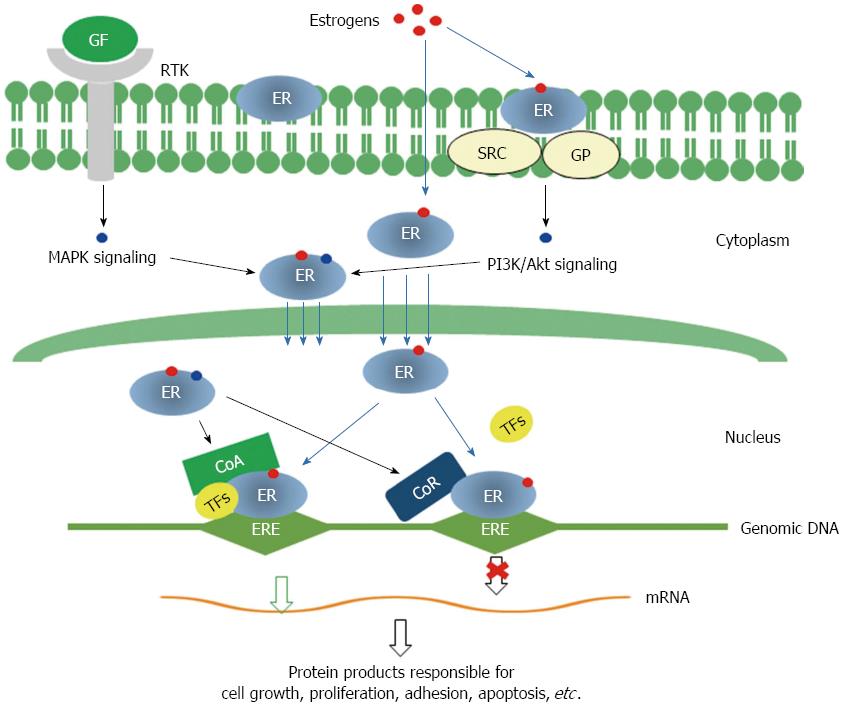
Figure 2 Molecular mechanisms for the functions of estrogen receptors.
Genomic pathway: Estrogen binding leads to dimerization of ERs, then ERs translocate into nucleus and interact with transcriptional co-activators and/or co-repressor and bind to genomic DNA at specific sequences known as estrogen response elements (EREs) to activate or repress the transcription of specific genes. Non-genomic signaling pathway: Membrane ERs interact with SRC/G protein and activate PI3K/Akt signaling. Both MAPK signaling initiated by binding of growth factors to receptor tyrosine kinases and PI3K/Akt signaling can modify cytosolic ERs, which may interact with other transcription factors and modulate the transcription of specific genes. GF: Growth factor; RTK: Receptor tyrosine kinase; GP: G proteins; CoA: Transcription co-activator; CoR: Transcription co-receptor; TFs: Transcription factors; MAPK: Mitogen activated protein kinase; PI3K: Phosphoinositide 3 kinase; ERs: Estrogen receptors.
- Citation: Rahman MSU, Cao J. Estrogen receptors in gastric cancer: Advances and perspectives. World J Gastroenterol 2016; 22(8): 2475-2482
- URL: https://www.wjgnet.com/1007-9327/full/v22/i8/2475.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i8.2475