Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Dec 28, 2014; 20(48): 18249-18259
Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18249
Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18249
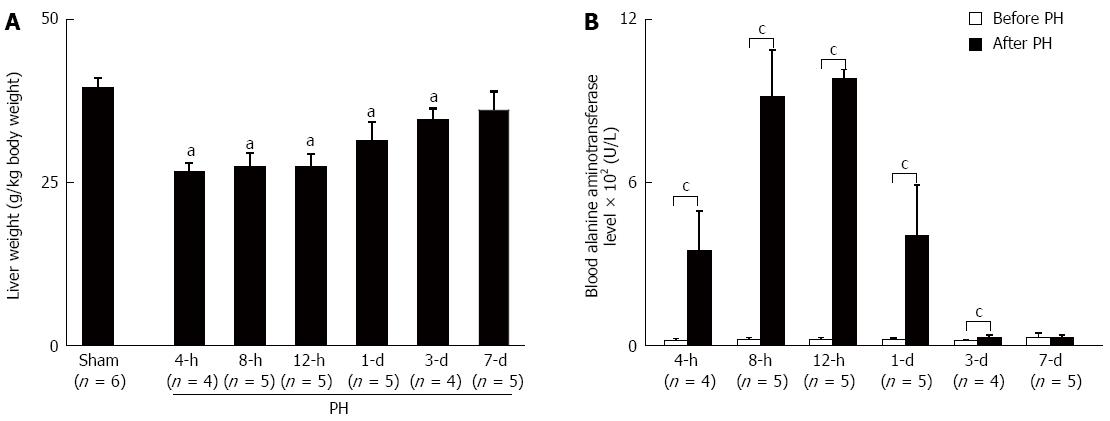
Figure 1 Seven days after the operation, the liver weight and function had recovered.
A: Liver weight; B: Liver function. All of the values shown are the mean ± SD. aP < 0.05 vs the Sham group using ANOVA; cP < 0.05 vs the before PH by 2-tailed Student t test. PH: Partial hepatectomy.
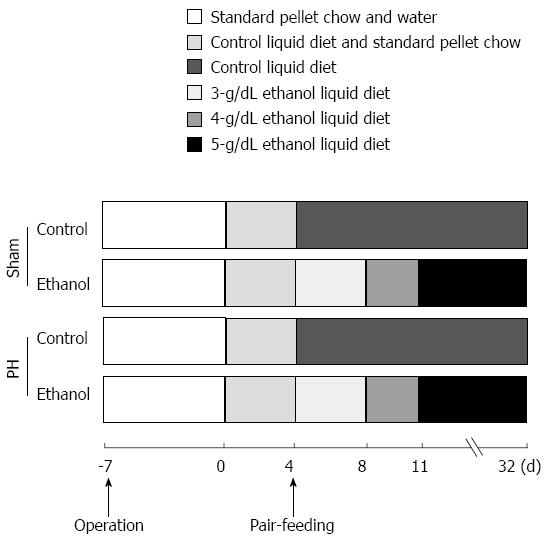
Figure 2 Chronic ethanol treatment protocol.
PH: Partial hepatectomy.
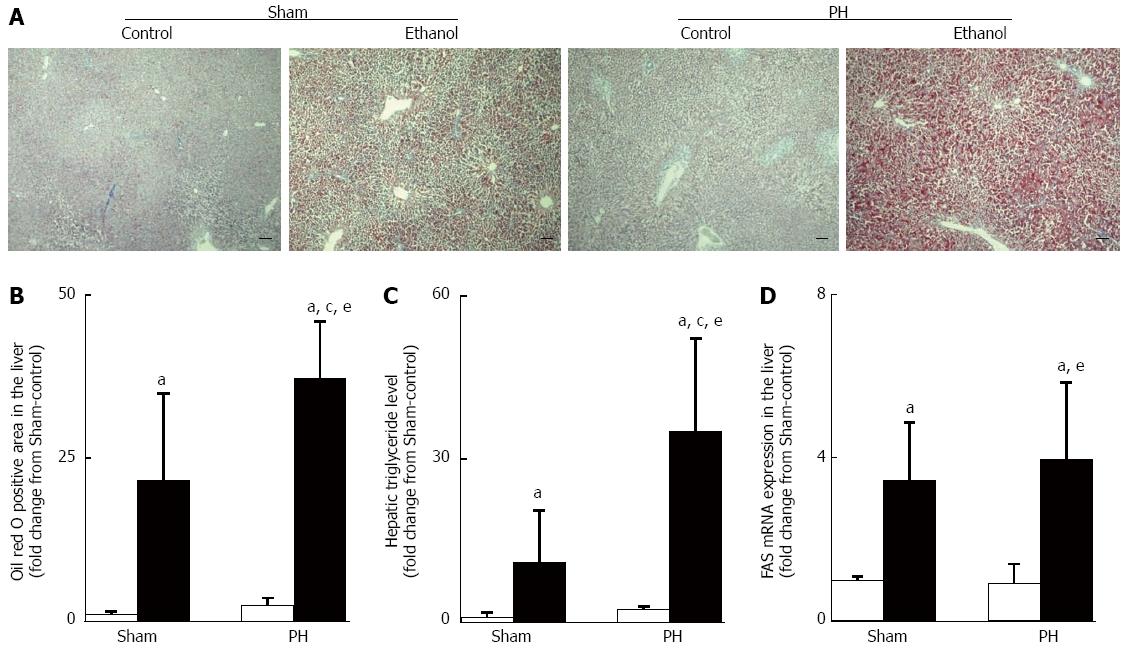
Figure 3 Ethanol-induced hepatic steatosis.
A: Representative examples of Oil red O stains of liver sections, scale bars = 100 μm; B: Oil red O positively stained area; C: Hepatic triglyceride levels; D: Fas mRNA expression. The open bar corresponds to the rats fed the control liquid diet, and the closed bar corresponds to the rats fed the ethanol liquid diet. All of the values shown are the mean ± SD, with 5 rats in each group, except for the hepatic triglyceride level values, which represent the mean ± SD of 4 rats per group. aP < 0.05 vs the Sham-control group, cP < 0.05 vs the Sham-ethanol group, eP < 0.05 vs the PH-control group using ANOVA. PH: Partial hepatectomy.
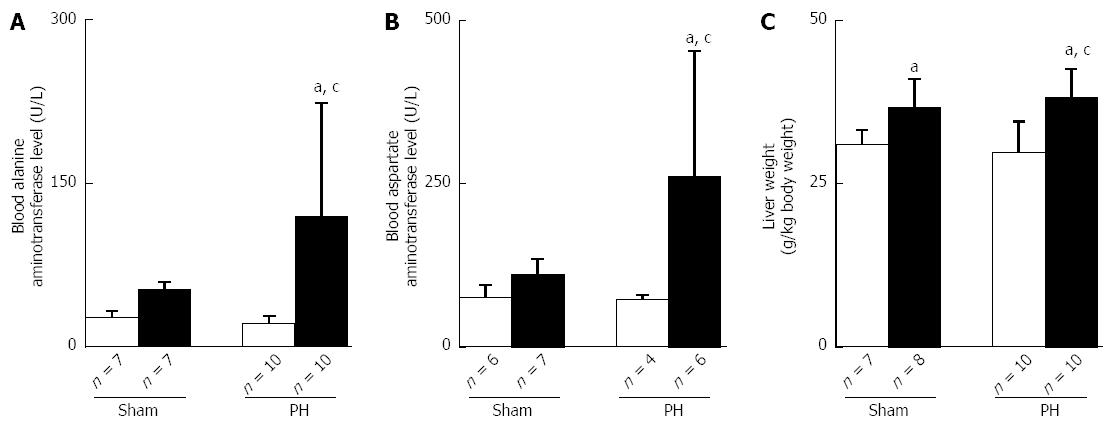
Figure 4 Ethanol-induced liver dysfunction and the increases in liver weight.
A: ALT; B: AST; C: liver weight. The open bar corresponds to the rats fed the control liquid diet, and the closed bar corresponds to the rats fed the ethanol liquid diet. All of the values shown are the mean ± SD. aP < 0.05 vs the Sham-control group,cP < 0.05 vs the PH-control group using ANOVA. PH: Partial hepatectomy.
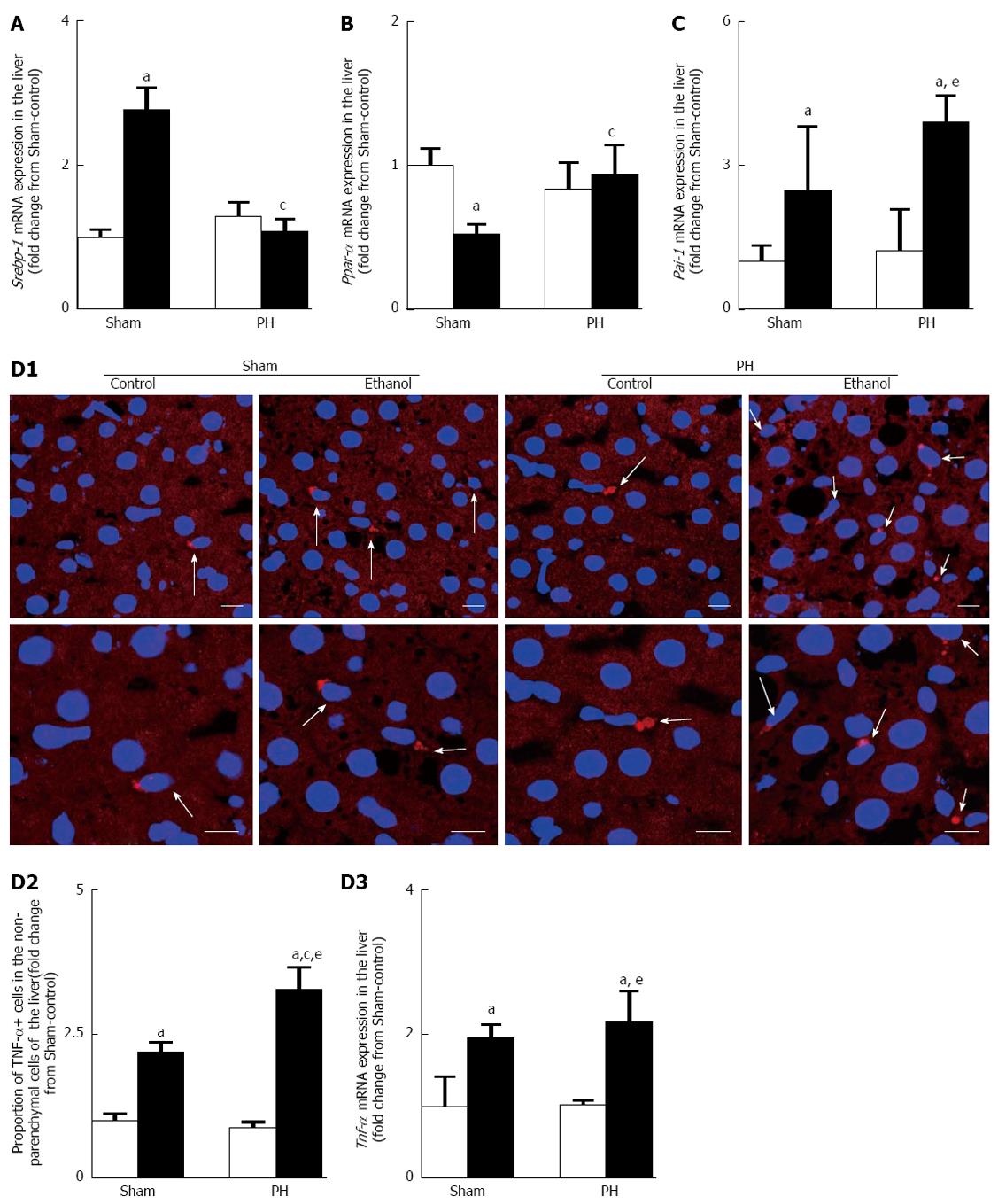
Figure 5 Ethanol modified the gene expression levels of lipid metabolism-associated enzyme regulators and up-regulated the gene expression of the mediators that alter lipid metabolism.
A: Srebp-1 mRNA expression; B: Ppar-α mRNA expression; C: Pai-1 mRNA expression; D: TNF-α overproduction (D1: Examples of immunohistochemical images of TNF-α positive cells in liver sections. A portion of each upper panel is shown under higher magnification in its corresponding lower panel, scale bars = 10 μm; D2: Quantitative immunohistochemical analysis of the TNF-α positive cells; D3: Tnf-α mRNA expression). The open bar corresponds to the rats fed the control liquid diet, and the closed bar corresponds to the rats fed the ethanol liquid diet. All of the values are shown as the mean ± SD, with 4 rats in each group, except for the quantitative immunohistochemical values for TNF-α positive cell, which represent the mean ± SD of 5 rats per group. aP < 0.05 vs the Sham-control group, cP < 0.05 vs the Sham-ethanol group, eP < 0.05 vs the PH-control group using ANOVA. PH: Partial hepatectomy; TNF: Tumor necrosis factor; SREBP: Sterol regulatory element binding protein; PPAR: Peroxisome proliferator-activated receptor; PAI: Plasminogen activator.
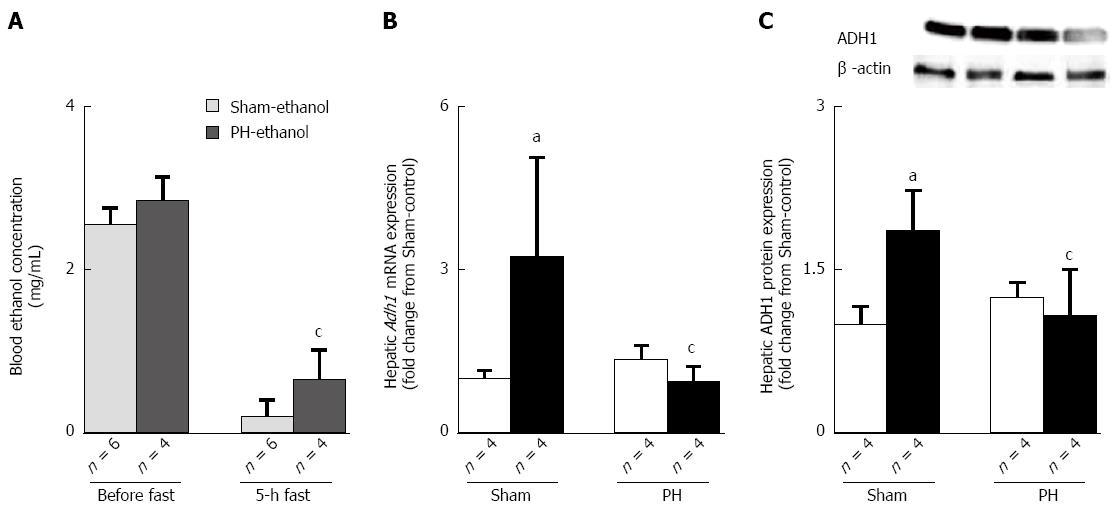
Figure 6 Rats had an alcohol dehydrogenase ethanol elimination delay after their liver resections.
A: The blood ethanol concentrations before and after a 5-h fast; B: ADH1 mRNA expression; C: ADH1 protein expression. The open bar corresponds to the rats fed the control liquid diet, and the closed bar corresponds to the rats fed the ethanol liquid diet. All of the values are shown as the mean ± SD. aP < 0.05 vs the Sham-control group, cP < 0.05 vs the Sham-ethanol group by ANOVA. PH: Partial hepatectomy.
- Citation: Liu X, Hakucho A, Liu J, Fujimiya T. Delayed ethanol elimination and enhanced susceptibility to ethanol-induced hepatosteatosis after liver resection. World J Gastroenterol 2014; 20(48): 18249-18259
- URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18249.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18249