Copyright
©2010 Baishideng.
World J Gastroenterol. Mar 21, 2010; 16(11): 1349-1357
Published online Mar 21, 2010. doi: 10.3748/wjg.v16.i11.1349
Published online Mar 21, 2010. doi: 10.3748/wjg.v16.i11.1349
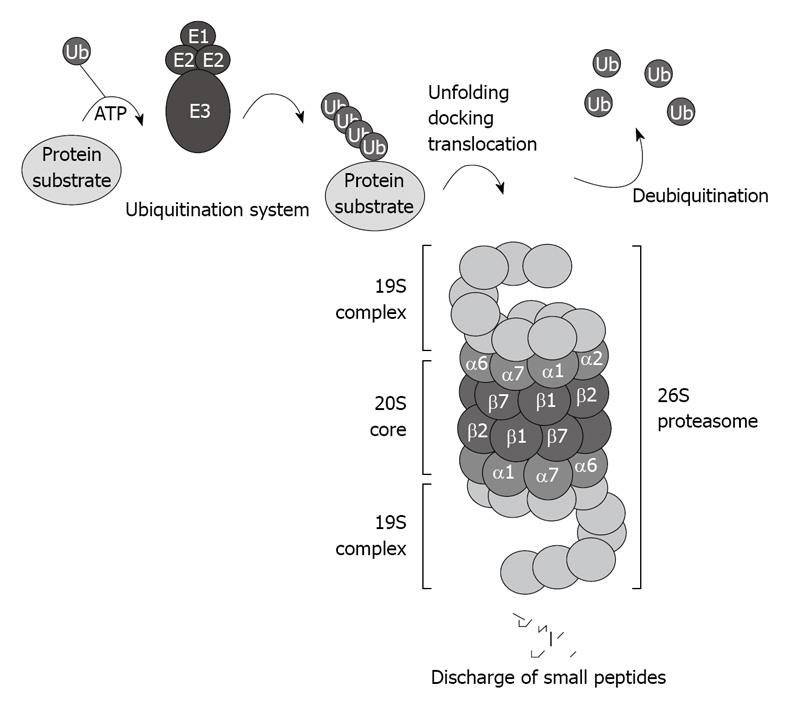
Figure 1 Schematic illustration of the ubiquitin proteasome pathway.
The 26S proteasome consists of the 20S core capped with the 19S regulatory complexes that recognize ubiquitinated protein substrates designated for proteolysis.
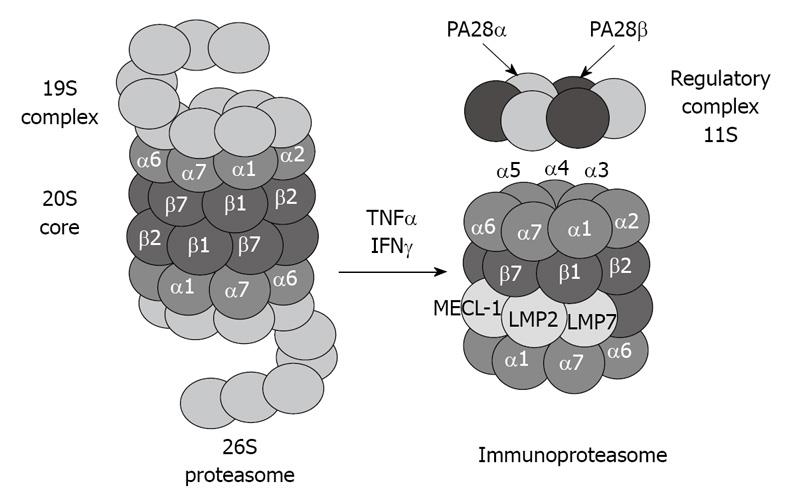
Figure 2 Tumor necrosis factor (TNF) α and interferon (IFN) γ induce formation of immunoproteasome subunits.
LMP7, LMP2 and MECL-1 subunits replace the constitutive catalytic subunits β5, β1 and β2, respectively, which shift the catalytic properties of the proteasome to generate MHC-I-binding peptides.
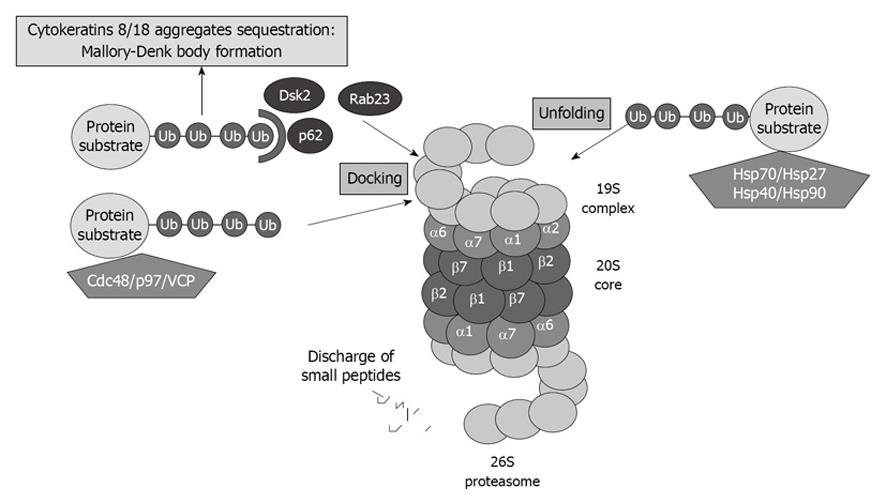
Figure 3 Proteasome interacting proteins (PIPs) involved in unfolding and docking of protein substrates designated for degradation by the proteasome.
VCP: Valosin-containing protein.
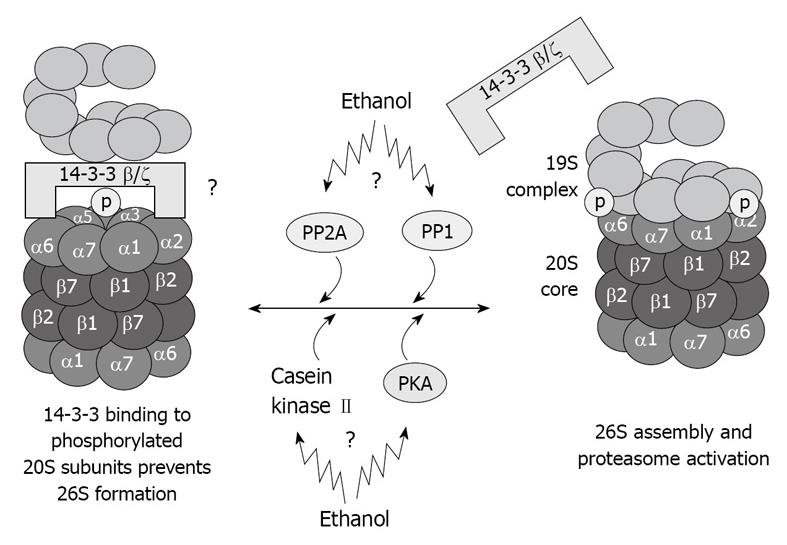
Figure 4 Illustration of hypothetical chronic ethanol feeding effects in 26S dysfunction.
As a result of phosphorylation deregulation, 20S and 19S binding is altered, which causes 26S dysfunction. Total dephosphorylation of proteasome subunits by phosphatases is prevented by 14-3-3 protein sequestration of the 20S proteasome.
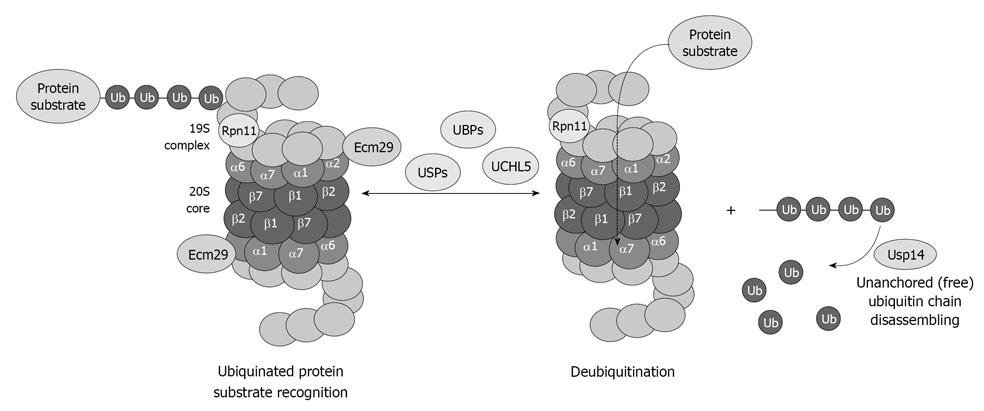
Figure 5 Diagram showing the important role of the deubiquitinases in the process of proteasome function and removal of ubiquitinated protein substrates, as well as recycling of free ubiquitin.
- Citation: Bardag-Gorce F. Effects of ethanol on the proteasome interacting proteins. World J Gastroenterol 2010; 16(11): 1349-1357
- URL: https://www.wjgnet.com/1007-9327/full/v16/i11/1349.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i11.1349