Copyright
©The Author(s) 2005.
World J Gastroenterol. Aug 7, 2005; 11(29): 4511-4518
Published online Aug 7, 2005. doi: 10.3748/wjg.v11.i29.4511
Published online Aug 7, 2005. doi: 10.3748/wjg.v11.i29.4511
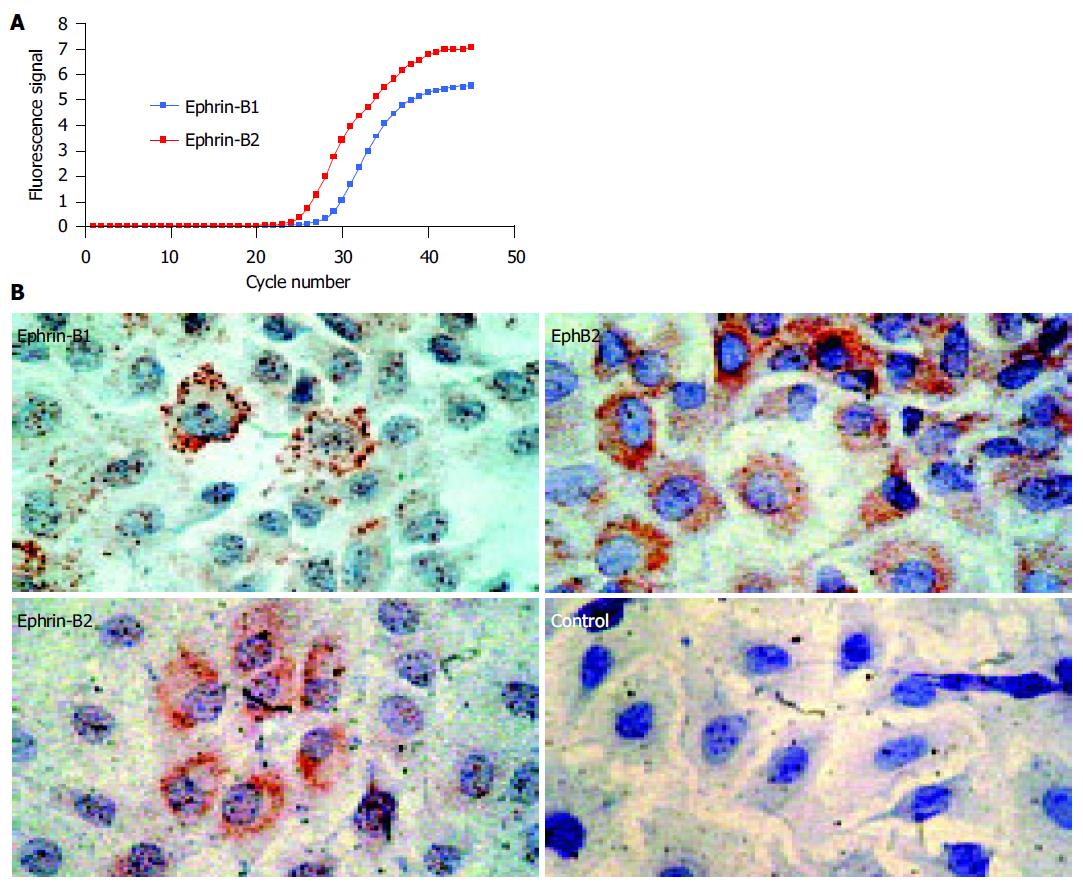
Figure 1 Rat IEC-6 intestinal epithelial cells express ephrin-B ligands on the mRNA and protein level.
A: Real-time RT-PCR shows expression of ephrin-B1 and -B2 mRNA. As the curve of ephrin-B2 demonstrates lower CT-values, the amount of ephrin-B2 mRNA starting material was higher than the amount of ephrin-B1 mRNA. The values were normalized by detection of the 18S rRNA (data not shown); B: IEC-6 cells coexpress ephrin-B1/2 and the corresponding EphB2 receptor on the protein level. IEC-6 cells may therefore be able to interchange cell-cell signals via EphB2/ephrin-B1/2 contacts. The coexpression of the EphB2 receptors and ephrin-B1/2 ligands is in accordance with previous data on human adult intestinal tissue in these cells[11].
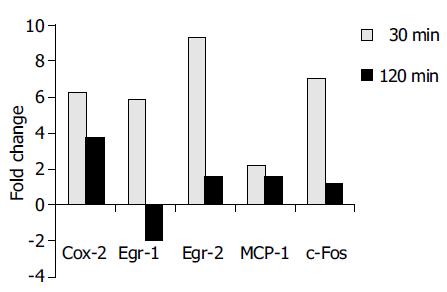
Figure 2 Real-time RT-PCR analysis confirmed the differential expression of COX-2, Egr-1, Egr-2, MCP-1, and c-Fos in accordance to Affymetrix® array data.
The results are indicated as positive ( = upregulation) or negative ( = downregulation) fold changes of mRNA amount compared to the control (t = 0 min, values normalized to 18S rRNA).
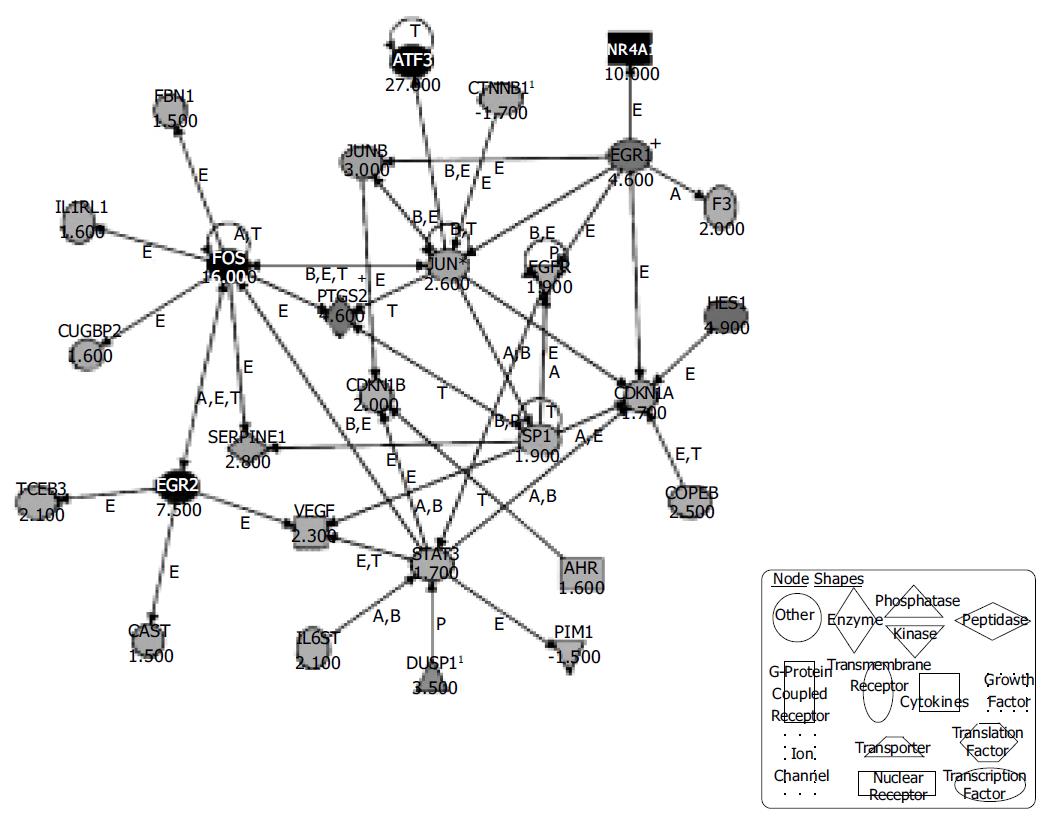
Figure 3 Ingenuity® Pathways Analysis reveals a possible network of genes activated in IEC-6 cells upon induction of the ephrin-B pathway.
Three hundred and thirty-one regulated genes with changed -P values lower than 0.003 and higher than 0.997 were selected and entered the Ingenuity® pathway analysis. The displayed network features only genes with functional interactions. Many of those genes are associated with cellular growth, proliferation and control of cell death. Arrows pointing from one gene to another indicate that one causes activation of the other one (includes any direct interaction: e.g. binding, phosphorylation, dephosphorylation, etc.). For ligands/receptors arrows pointing from a ligand to a receptor signify that the ligand binds the receptor and subsequently leads to activation of the receptor. The respective fold-changes are given below the gene symbols. Further abbreviations are: A, Activation/deactivation; B, Binding; E, Expression; M, Biochemical Modification; P, Phosphorylation/dephosphorylation; T, Transcription; 1Duplicate-user input gene that had duplicate identifiers in the dataset file mapping to a single gene in the Ingenuity® Pathways Knowledge Base. + Indicates there are other networks from the analysis that contain this gene. Gene names are: AHR: Aryl hydrocarbon receptor; ATF3: Activating transcription factor 3; CAST: Calpastatin; CDKN1A: Cyclin-dependent kinase inhibitor 1A; CDKN1B: Cyclin-dependent kinase inhibitor 1B; COPEB: Core promoter element binding protein; CTNNB1: Catenin (cadherin-associated protein), beta 1; CUGBP2: UG triplet repeat, RNA-binding protein 2; DUSP1: Dual specificity phosphatase 1; EGFR: Epidermal growth factor receptor; EGR1: Early growth response 1; EGR2: Early growth response 2; F3: Coagulation factor 3; FBN1: Fibrillin-1; FOS: v-fos FBJ murine osteosarcoma viral oncogene homolog; HES1: Hairy and enhancer of split 1 (Drosophila); IL1RL1: Interleukin 1 receptor-like 1; IL6ST: Interleukin 6 signal transducer; JUN: v-jun sarcoma virus 17 oncogene homolog (avian); JUNB: Jun-B oncogene; NR4A1: Nuclear receptor subfamily 4, group A, member 1; PIM1: Proviral integration site 1; PTGS2: Prostaglandin-endoperoxide synthase 2; SERPINE1: Serine (or cysteine) proteinase inhibitor, member 1; SP1: Sp1 transcription factor; STAT3: Signal transducer and activator of transcription 3; TCEB3: Transcription elongation factor B (SIII), polypeptide 3; VEGF: Vascular endothelial growth factor.
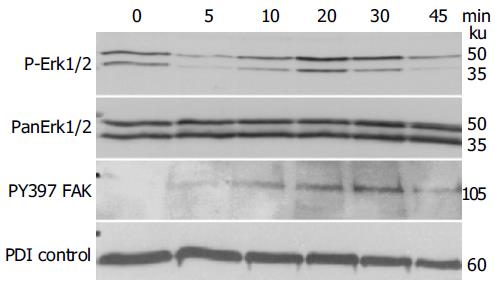
Figure 4 Stimulating the ephrin-B reverse signaling pathway in IEC-6 with recombinant EphB1-Fc induces temporary activation of Erk1/2 map kinase stress pathways and FAK phosphorylation, demonstrated by immunoblotting.
PDI served as a loading control.
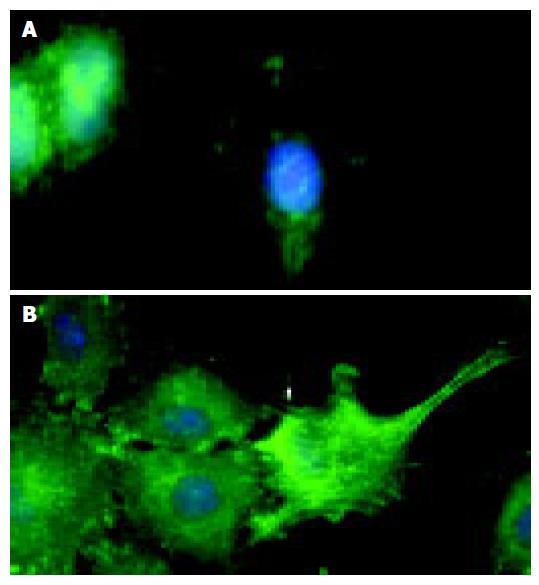
Figure 5 Immunofluorescence microscopy demonstrates that compared to the control (A), stimulation of IEC-6 cells with recombinant EphB1-Fc leads to induction of active pY397-FAK in neighboring cells most prominently at the wound edge (B).
- Citation: Hafner C, Meyer S, Hagen I, Becker B, Roesch A, Landthaler M, Vogt T. Ephrin-B reverse signaling induces expression of wound healing associated genes in IEC-6 intestinal epithelial cells. World J Gastroenterol 2005; 11(29): 4511-4518
- URL: https://www.wjgnet.com/1007-9327/full/v11/i29/4511.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i29.4511