Published online Jun 27, 2014. doi: 10.4254/wjh.v6.i6.435
Revised: March 1, 2014
Accepted: May 16, 2014
Published online: June 27, 2014
AIM: To investigate genetic susceptibility in Indian subjects with non-alcoholic fatty liver disease (NAFLD) by performing a pooled genetic study.
METHODS: Study subjects (n = 306) were recruited and categorized into NAFLD and control groups based on ultrasound findings of fatty infiltration. Of the 306 individuals, 156 individuals had fatty infiltration and thus comprised the NAFLD group. One hundred and fifty (n = 150) individuals were normal, without fatty infiltration of the liver, comprising the control group. Blood samples, demographic and anthropometric data from the individuals were collected after obtaining informed consent. Anthropometric data, blood glucose, lipids and liver function tests were estimated using standard methods. Genome wide association studies done to date on NAFLD were identified, 19 single nucleotide polymorphisms (SNPs) were selected from these studies that were reported to be significantly associated with NAFLD and genotyping was performed on the Sequenom platform. Student’s t test for continuous variables and χ2 test was applied to variant carriers from both groups. Required corrections were applied as multiple testing was done.
RESULTS The mean age of the control group was 39.78 ± 10.83 and the NAFLD group was 36.63 ± 8.20 years. The waist circumference of males and females in the control and NAFLD groups were 80.13 ± 10.35; 81.77 ± 13.65 and 94.09 ± 10.53; 92.53 ± 8.27 cms respectively. The mean triglyceride and alanine transaminase (ALT) levels in the control and NAFLD groups were 135.18 ± 7.77 mg/dL; 25.39 ± 14.73 IU/L and 184.40 ± 84.31 mg/dL; 110.20 ± 67.05 IU/L respectively. When χ2 test was applied to the number of individuals carrying the variant risk alleles between the control and NAFLD group, a significant association was seen between rs738409 of the patatin-like phospholipase domain containing 3 (PNPLA3) gene (P = 0.001), rs2073080 of the PARVB gene (P = 0.02), rs2143571 of SAMM50 gene (P = 0.05) and rs6487679 of the pregnancy zone protein (PZP) gene (P = 0.01) with the disease. Variant single nucleotide polymorphisms (SNPs) in NCAN and PNPLA3 gene were associated with higher levels of ALT, whereas variant SNPs in APOC3, PNPLA3, EFCAB4B and COL13A1 were associated with high triglyceride levels. Apart from the above associations, rs2073080, rs343062 and rs6591182 were significantly associated with high BMI; rs2854117 and rs738409 with high triglyceride levels; and rs2073080, rs2143571, rs2228603, rs6487679 and rs738409 with high ALT levels.
CONCLUSION: Pooled genetic analysis revealed an association of SNPs in PNPLA3, PARVB, SAMM50 and PZP genes with NAFLD. SNPs in NCAN and PNPLA3 gene were associated with higher levels of ALT, whereas variant SNPs in APOC3, PNPLA3, EFCAB4B and COL13A1 were associated with high triglyceride levels.
Core tip: Non-alcoholic fatty liver disease (NAFLD) describes a range of conditions caused by build-up of fat within liver cells in the absence of alcohol consumption. Although obesity, diabetes, age, hypertension and hypertriglyceridemia contribute to the disease, genetics also has an important role to play. Furthermore, in 26%-35% of patients, genetic component is believed to contribute to NAFLD. By identifying significant single nucleotide polymorphisms from genome wide association studies reported from different ethnic populations for NAFLD and performing a pooled genetic association study, this study has identified important genetic risks that could help in identifying individuals with susceptibility at an early stage, thus aiding in better management of the disease.
- Citation: Kanth VVR, Sasikala M, Rao PN, Steffie Avanthi U, Rao KR, Nageshwar Reddy D. Pooled genetic analysis in ultrasound measured non-alcoholic fatty liver disease in Indian subjects: A pilot study. World J Hepatol 2014; 6(6): 435-442
- URL: https://www.wjgnet.com/1948-5182/full/v6/i6/435.htm
- DOI: https://dx.doi.org/10.4254/wjh.v6.i6.435
Non-alcoholic fatty liver disease (NAFLD) is a global epidemic, the incidence of which is reported to be as high as 25%-30% in different populations[1]. Differences in prevalence, clinical profile, histological severity and outcome of NAFLD in different ethnic groups suggest a genetic contribution; and NAFLD in 26%-35% of patients is believed to be contributed by genetic component[2,3]. In recent years, genetic heritability has been a major focus of research, although changing dietary habits and modifying life style have been demonstrated to benefit patients with hepatic steatosis[4]. Genome wide association studies (GWAS) from different ethnic populations revealed a strong association of PNPLA3 variant[3,5], apart from few other variants[6-8], and an independent study identified APOC3 variants associated with higher triglyceride levels and risk of NAFLD in migrant Indians[9].
A recent GWAS[6] of hepatic steatosis revealed loci in or near the neurocan (NCAN), glucokinase regulatory protein, lysophospholipase-like protein 1 and protein phosphatase 1, regulatory subunit 3B (PPP1R3B) genes that have associations with glycemic traits, serum lipid levels, hepatic steatosis, hepatic inflammation/fibrosis, or a combination of these. Specific genotypic information in the form of single nucleotide polymorphisms (SNPs) which confer susceptibility for an individual have to be identified so that early preventive measures can be initiated, especially in children and adolescents. Patatin-like phospholipase domain containing 3 (PNPLA3) missense variant was studied and compared with MR spectroscopy for predicting NAFLD[4]. However, since it is now known that multiple SNPs are associated with the disease, identifying other susceptibility SNPs apart from PNPLA3 would enhance the predictive capability.
The prevalence of NAFLD in the Indian population is estimated to be around 25%-30%[10-16]. In addition, the prevalence of hepatic steatosis in non-obese (lean NAFLD) was shown to range between 11%-31.7% according to a recent study[17]. Increase in the incidence of obesity, metabolic syndrome and the presence of lean non-alcoholic steatohepatitis (NASH) in the Indian population warrants genetic susceptibility studies in Indian NAFLD subjects. In this preliminary pilot study, we selected SNPs (Table 1) from already reported GWAS across different populations and genotyped the same in Indian subjects.
| SNP No | rsID | Risk allele | Associated gene | Associated with | Ref. |
| 1 | rs738409 | G | PNPLA3 | Hepatic steatosis | [6] |
| 2 | rs4240624 | A | PPP1R3B | Hepatic steatosis | [6] |
| 3 | rs2228603 | T | NCAN | Hepatic steatosis | [6] |
| 4 | rs780094 | A | GCKR | Hepatic steatosis | [6] |
| 5 | rs12137855 | C | LYPLAL1 | Hepatic steatosis | [6] |
| 6 | rs2645424 | C | FDFT1 | NAFLD activity score | [8] |
| 7 | rs343062 | T | - | Degree of fibrosis | [8] |
| 8 | rs1227756 | G | COL13A1 | Lobular inflammation | [8] |
| 9 | rs6591182 | G | - | Lobular inflammation | [8] |
| 10 | rs887304 | A | EFCAB4B | Lobular inflammation | [8] |
| 11 | rs2499604 | A | Intronic ZP4-TRNAP23P | Serum levels of alanine aminotransferase | [8] |
| 12 | rs6487679 | C | PZP | Serum levels of alanine aminotransferase | [8] |
| 13 | rs1421201 | C | - | Serum levels of alanine aminotransferase | [8] |
| 14 | rs2710833 | T | - | Serum levels of alanine aminotransferase | [8] |
| 15 | rs2854116 | A | APOC3 | Hypertriglyceridemia in Asians | [9] |
| 16 | rs2854117 | G | APOC3 | Hypertriglyceridemia in Asians | [9] |
| 17 | rs2143571 | A | SAMM50 | NAFLD | [7] |
| 18 | rs2073080 | T | PARVB | NAFLD | [7] |
| 19 | rs1390096 | A | HS3ST1-HSP90AB2P | NAFLD | [7] |
| 20 | rs11206226 | A | YIPF1 | NAFLD | [7] |
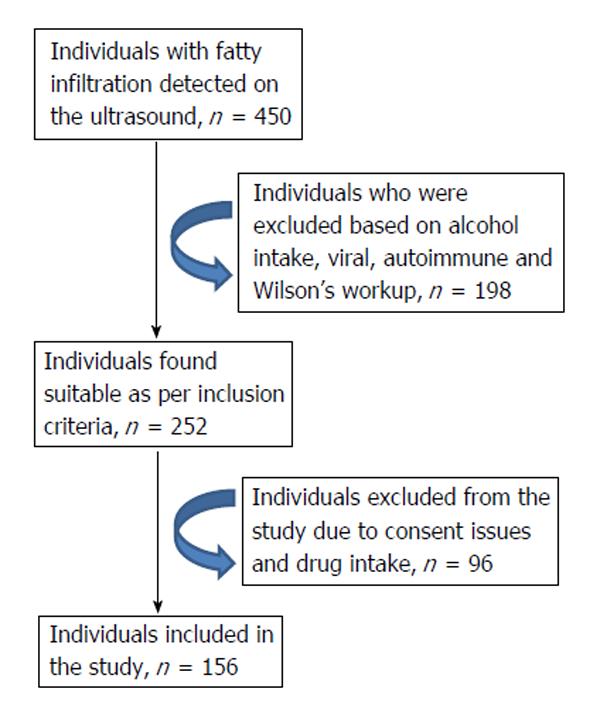
A total of 450 individuals with fatty infiltration were recruited for the study during 2011-2012 (1 year) from hepatology clinics of the hospital. As shown (Figure 1), 156 individuals were found to be eligible for the pooled genetic analysis. Statistical power analysis was not used to compute the sample size as this is a pilot study. Although liver biopsy is considered to be the gold standard for identifying NAFLD and NASH, lack of indication for asymptomatic individuals, the costs involved, risk of complications and ethical concerns limit its use in these types of studies. Therefore, subjects were recruited based on ultrasound findings of hepatic steatosis as per earlier reports[18,19]. Healthy subjects (n = 150) from the institute who volunteered to be part of the study were recruited as controls based on the sole criteria of the absence of fatty liver on ultrasonography and normal alanine transaminase (ALT) levels. Written informed consent was obtained from each individual. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Committee. Demographic and anthropometric details [height, weight, magnetic resonance imaging (BMI) and waist circumference] were collected in a structured pro forma. Whole blood (5 mL) was collected in pre coated EDTA containers from the study group and stored at -20 °C until further analysis. Biochemical investigations like ALT, viral markers and lipid profiles were estimated as per standard methods.
Individuals with BMI less than 18.5 kg/m2 were defined as underweight; 18.5-22.9 kg/m2 were defined as normal and BMI more than 23 kg/m2 were defined as obese. Lean NAFLD was defined as hepatic steatosis in individuals with normal BMI (< 22.9 kg/m2) according to Asian standards[20]; likewise hypertriglyceridemia (greater than 150 mg/dL), low levels of high density lipoprotein (HDL less than 40 mg/dL in males and 50 mg/dL in females), hypertension (greater than 130/85 systolic and diastolic blood pressure level in mmHg or on anti hypertensive drugs) and high fasting glucose levels (greater than 100 mg/dL of fasting blood sugar levels) were considered as cut offs. The cut off for waist circumference was > 90 cm and > 80 cm in males and females respectively, as per Asian standards[21]. A cut off of 30 IU/L was considered for ALT[22].
DNA was isolated from blood using standard protocols. The concentration and integrity of DNA was measured with NanoDrop 1000 spectrophotometer (Thermo Scientific, USA) and agarose gel electrophoresis respectively. The DNA with 260/280 ratios between 1.8-2.0 and agarose gel image showing a high molecular weight intact DNA band were included for further genotyping analysis. The samples were genotyped for the 19 SNPs on the Sequenom platform (Sequenom®, San Diego, CA, United States) using the manufacturer’s protocol. Primers for one SNP (rs2854116) could not be designed because of proximal SNPs present very near to the target SNP and so was not included in the study. The raw data files generated by Sequenom MassARRAY were analyzed for the intensity peaks of calibrant to ascertain the quality of the data. An overall call rate of > 95% was maintained. Five percent of the samples were duplicated across the plate, their genotypes compared and they had 100% concordance. Negative controls (master mix without DNA) were also included.
Correlation of demographic and anthropometric phenotypes, like BMI, waist circumference, liver enzymes (ALT) and triglyceride levels, to the genotype was done to identify significant risk factors.
The data collected was edited for consistency and completeness and entered into MS-Excel for further analysis. Patient characteristics were compared using Student’s t test for continuous variables and proportion test for categorical variables. χ2 test was used on the number of variant carriers in the control and NAFLD groups for identifying SNPs associated with NAFLD. To correct for multiple comparison testing, the Benjamini and Hochberg false discovery rate correction[23] was applied to “P values”. All SNPs were divided into risk and non-risk groups and 2X2 contingency tables were prepared to estimate odds ratio for all variables like age, gender, BMI, ALT levels etc. Multiple logistic regression was used to identify independent predictor variables for NAFLD. The data was analyzed using Statistical Package for Social Sciences (SPSS Version 17). In this study, a P value ≤ 0.05 was considered statistically significant. Haplotype analysis was carried out using software[24]. An excel sheet was prepared as per instructions with “0” representing wild type allele and “1” representing heterozygous or mutant variants.
The clinical characteristics, such as age, waist circumference, BMI, triglyceride and ALT levels, of the groups are presented in Table 2. Categorization of the study population yielded two groups based on ultrasonographic detection of hepatic steatosis in the liver, namely the NAFLD (n = 156) and control group (n = 150). The inclusion of individuals in the control group was based on the absence of hepatic steatosis and retrospectively it was seen that few of the individuals in the control group were obese (BMI > 23 kg/m2). So the group was divided based on BMI and a comparison of both clinical characteristics and the genotype was made (data not shown) between the normal and obese controls. Such an analysis did not show any significant differences between the normal and obese control group with respect to the genotype. However, the waist circumference in males (P = 0.0001) and females (P = 0.02) and the triglyceride levels (P = 0.0057) were high in the obese controls, apart from BMI (P = 0.0001), and the difference in all the other characteristics studied was statistically not significant.
| Parameter | Controls | Patients | P value |
| (n = 150) | (n = 156) | ||
| Mean ± SD | 39.78 ± 10.83 | 36.63 ± 8.20 | 0.004 |
| Age, yr | 18-63 | 19-62 | |
| Range | |||
| Males | 110 (73.33%) | 138 (88.46%) | - |
| Females | 40 (26.66%) | 18 (11.53%) | - |
| Waist circumference | |||
| Males | 80.13 ± 10.35 | 94.09 ± 10.53 | 0.0001 |
| Females | 81.77 ± 13.65 | 92.53 ± 8.27 | 0.01 |
| BMI (kg/m2) | 24.04 ± 7.77 | 27 ± 5.86 | 0.001 |
| Triglycerides (mg/dL) | 135.18 ± 7.77 | 184.40 ± 84.31 | 0.0001 |
| HDL (mg/dL) | 41.86 ± 9.70 | 39.56 ± 13.02 | 0.2923 |
| ALT (IU/L) | 25.39 ± 14.73 | 110.20 ± 67.05 | 0.0001 |
| AST (IU/L) | 25.99 ± 8.46 | 69.14 ± 37.77 | 0.0001 |
| Hypertensives | 4 (2.6%) | 18 (11.53%) | - |
| Diabetics | 19 (12.66%) | 27 (17.3%) | - |
When an analysis was done between the control and NAFLD group, a significant difference in clinical characteristics was noted in BMI (P = 0.001), waist circumference of males (P = 0.0001) and females (P = 0.01), high triglyceride levels (P = 0.0001) and ALT (P = 0.0001) levels in the NAFLD group.
In the single allelic analysis, tests for associations between NAFLD and the SNPs revealed that variants in PARVB, SAMM50, NCAN, intronic SNP (rs2499604), APOC3, pregnancy zone protein (PZP) and PNPLA3 genes were associated with NAFLD; however, after correction for multiple testing was applied, only variants in PARVB, SAMM50, PZP and PNPLA3 were significant (Table 3).
| SNP- | Allele frequency controls (n = 150) | Allele frequency patients (n = 156) | χ2 | P value | Corrected P value1 | OR | 95%CI | ||
| gene name | Major | Minor | Major | Minor | lower-upper | ||||
| rs1227756 | 0.49 | 0.51 | 0.44 | 0.56 | 0.02 | 0.88 | 0.93 | 1.04 | 0.54-2.03 |
| COL13A1 | |||||||||
| rs12137855 | 0.77 | 0.23 | 0.73 | 0.27 | 0.41 | 0.51 | 0.62 | 1.20 | 0.68-2.12 |
| LYPLAL1 | |||||||||
| rs1390096 | 0.68 | 0.32 | 0.66 | 0.34 | 0.007 | 0.93 | 0.93 | 0.97 | 0.55-1.70 |
| HS3ST1-HSP | |||||||||
| rs1421201 | 0.88 | 0.12 | 0.85 | 0.15 | 1.60 | 0.20 | 0.36 | 1.55 | 0.78-3.10 |
| intronic | |||||||||
| rs2073080 | 0.81 | 0.19 | 0.69 | 0.31 | 8.42 | 0.003 | 0.02 | 2.36 | 1.31-4.22 |
| PARVB | |||||||||
| rs2143571 | 0.80 | 0.20 | 0.69 | 0.31 | 6.25 | 0.01 | 0.05 | 2.07 | 1.16-3.69 |
| SAMM50 | |||||||||
| rs2228603 | 0.97 | 0.03 | 0.92 | 0.08 | 4.09 | 0.04 | 0.12 | 3.29 | 1.10-9.84 |
| NCAN | |||||||||
| rs2499604 | 0.53 | 0.47 | 0.60 | 0.40 | 3.76 | 0.05 | 0.13 | 1.92 | 0.98-3.75 |
| intronic | |||||||||
| rs2645424 | 0.5 | 0.50 | 0.56 | 0.44 | 1.21 | 0.27 | 0.40 | 0.69 | 0.36-1.32 |
| FDFT1 | |||||||||
| rs2710833 | 0.60 | 0.40 | 0.66 | 0.34 | 3.09 | 0.07 | 0.17 | 0.56 | 0.30-1.07 |
| intronic | |||||||||
| rs2854117 | 0.58 | 0.42 | 0.55 | 0.45 | 4.24 | 0.03 | 0.12 | 1.83 | 1.02-3.28 |
| APOC3 | |||||||||
| rs343062 | 0.55 | 0.45 | 0.52 | 0.48 | 1.96 | 0.16 | 0.32 | 1.53 | 0.84-2.80 |
| intronic | |||||||||
| rs4240624 | 0.94 | 0.06 | 0.91 | 0.09 | 1.21 | 0.27 | 0.40 | 1.56 | 0.69-3.52 |
| PPP1R3B | |||||||||
| rs6487679 | 0.89 | 0.11 | 0.75 | 0.25 | 9.65 | 0.001 | 0.01 | 2.81 | 1.44-5.48 |
| PZP | |||||||||
| rs6591182 | 0.63 | 0.37 | 0.59 | 0.41 | 0.99 | 0.31 | 0.44 | 1.39 | 0.72-2.67 |
| intronic | |||||||||
| rs738409 | 0.92 | 0.08 | 0.60 | 0.40 | 46.37 | 0.0001 | 0.001 | 12.66 | 5.45-29.38 |
| PNPLA3 | |||||||||
| rs780094 | 0.76 | 0.24 | 0.74 | 0.26 | 0.48 | 0.48 | 0.62 | 1.22 | 0.69-2.15 |
| GCKR | |||||||||
| rs887304 | 0.82 | 0.18 | 0.83 | 0.17 | 0.19 | 0.65 | 0.73 | 0.87 | 0.47-1.59 |
| EFCAB4B | |||||||||
| rs11206226 | 1.00 | 0.00 | 1.00 | 0.00 | - | - | - | - | |
| YIPF1 | |||||||||
To identify SNPs which may be associated with clinical traits like triglyceride and ALT levels but not necessarily to the disease, the individuals in the study group were divided into two groups, namely individuals with normal and those with high levels of the mentioned clinical traits irrespective of the disease status. Such an effort identified significant SNPs which are likely to be associated with clinical traits. SNPs in NCAN (P = 0.04) and PNPLA3 (P = 0.001) were significantly associated with high ALT levels and SNPs in APOC3 (P = 0.01), PNPLA3 (P = 0.05), EFCAB4B (P = 0.04) and COL13A1 (P = 0.02) genes were significantly associated with high triglyceride levels.
Among the various characteristics like age, BMI and the SNPs that were studied, rs2073080 in PARVB, rs343062 (intronic) and rs6591182 (intronic) were significantly associated with higher odds of obese individuals with NAFLD. Likewise, SNPs in various genes studied were associated with clinical parameters like ALT and triglyceride levels (Table 4).
| Variable | SNP-gene | OR | P value | 95%CI |
| lower-upper | ||||
| BMI (obese and non-obese) | rs2073080-PARVB | 1.81 | 0.0470 | 1.00-3.25 |
| BMI (obese and non-obese) | rs343062-intronic | 2.25 | 0.0100 | 1.21-4.21 |
| BMI (obese and non-obese) | rs6591182-intronic | 2.05 | 0.0340 | 1.05-4.00 |
| TG (abnormal and normal) | rs2854117-APOC3 | 2.31 | 0.0040 | 1.29-4.13 |
| TG (abnormal and normal) | rs738409-PNPLA3 | 1.94 | 0.0170 | 1.12-3.37 |
| ALT (abnormal and normal) | rs2073080-PARVB | 1.92 | 0.0200 | 1.10-3.36 |
| ALT (abnormal and normal) | rs2143571-SAMM50 | 1.77 | 0.0200 | 1.10-3.36 |
| ALT(abnormal and normal) | rs2228603-NCAN | 3.23 | 0.0180 | 1.22-1.37 |
| ALT (abnormal and normal) | rs6487679-PZP | 1.92 | 0.0300 | 0.05-3.51 |
| ALT (abnormal and normal) | rs738409-PNPLA3 | 5.07 | 0.0001 | 0.03-10.92 |
Since PNPLA3, SAMM50 and PARVB are found on the same locus on chromosome 22, haplotype analysis was done for the 3 SNPs and it was noted that heterozygous or homozygous variants in these genes were overrepresented in the NAFLD group compared to the control group (8 in controls against 63 in the NAFLD group) (Table 5).
| Total counts | Haplotype | Number in controls | Number in patients |
| 138 | 000 | 90 | 48 |
| 31 | 001 | 6 | 25 |
| 10 | 010 | 10 | 0 |
| 4 | 011 | 2 | 2 |
| 4 | 100 | 2 | 2 |
| 48 | 110 | 32 | 16 |
| 71 | 111 | 8 | 63 |
Multiple logistic regression analysis was applied to the data to estimate the risk of an individual for NAFLD. The dependent variables were the NAFLD group and controls. The variables that were significant in the univariate analysis, namely age (less than 40 years), BMI, waist circumference, triglyceride levels, HDL, hypertension, diabetes and SNPs, were included for the multivariate analysis (Table 6).
| Variable | Regression | Standard | P value | OR | 95%CI | |
| coefficient | error | Lower | Upper | |||
| PZP | 0.880 | 0.415 | 0.034 | 2.41 | 1.05 | 5.44 |
| PNPLA3 | 2.289 | 0.480 | < 0.0001 | 9.86 | 3.85 | 25.29 |
| Triglyceride levels | 1.502 | 0.391 | 0.000 | 4.48 | 2.08 | 9.67 |
| Constant | -0.619 | |||||
The main objective of this study was to identify susceptibility SNPs for NAFLD in Indian subjects utilizing pooled genetic SNP data from various GWAS performed in different populations to date. Variants in SAMM50, PARVB, PZP and PNPLA3 genes were significantly associated with NAFLD, thus suggesting involvement of multiple loci in Indian NAFLD.
A significant association of PNPLA3 (rs738409) (P = 0.001) with NAFLD was observed in Indian subjects and is consistent with the genetic association of PNPLA3 in other populations, like Caucasians, European descent, Hispanics and Japanese[6-8]. Furthermore, this SNP was also significantly associated with higher ALT (P = 0.001) and triglyceride levels (P = 0.05), suggesting that individuals with the variant may be at higher risk for NAFLD. In addition to this SNP, PZP rs6487679 located on the 12th chromosome, demonstrated to have a role in clearance of transforming growth factor-beta from human plasma and hepatic fibrogenesis[25], was also significantly associated with NAFLD in Indian subjects. This finding corroborates with similar earlier findings in non-Hispanic Caucasians[8].
rs2073080 of the beta-parvin (PARVB) located on chromosome 22 that codes for a protein beta-parvin in humans[26] was significantly associated with the disease (P = 0.018) in the present study. Not much is known about the polymorphism, but in general the protein is believed to play a role in cytoskeleton organization and cell adhesion apart from having a role in tumor suppression (Entrez Gene: PARVB). The association of rs2143571 of the SAMM50 sharing the same locus on chromosome 22 as PNPLA3 and PARVB encoding sorting and assembly machinery component 50 homolog was also significantly associated with NAFLD in the Indian subjects[27]. This protein has a function in the assembly of beta-barrel proteins into the outer mitochondrial membrane. A recent genome wide scan[7,28] also identified similar SNPs in PNPLA3, SAMM50 and PARVB in the Japanese population which was significantly associated with NAFLD. Our results corroborate with this study, indicating that these 3 variants are commonly seen in an Asian population.
Apart from the promoter polymorphism of the APOC3 gene and variant in PNPLA3 gene, EFCAB4B and COL13A1 polymorphisms were identified as significantly associated with higher triglyceride levels. Polymorphisms in NCAN and PNPLA3 were associated with higher ALT levels. Although previous studies[6,7] reported an association of the above mentioned SNPs with NAFLD, their associations with triglycerides and ALT levels have been identified for the first time in Indian subjects.
When lean and obese controls were compared for significant differences in clinical characteristics and genotype, BMI between the groups was significantly different, with a higher BMI in the obese group as expected. The triglyceride levels were also significantly higher in the obese control group without hepatic steatosis compared to the lean controls; however, there were no significant differences in the genotype with respect to the 19 SNPs studied. Based on this, these individuals were found to be suitable to be included in the control group for further analyses.
The incidence of lean NAFLD in the present study (19.87%) is in agreement with an earlier study from North India[17] which had more or less a similar incidence (13.2%) and there was no significant difference in the incidence between the two groups (P = 0.08).
When odds were computed based on obese and non-obese status in the study group, PARVB and two intronic SNPs (rs343062 and rs6591182) were significantly associated with higher odds of NAFLD in the obese, suggesting that an obese individual with these variants is at a higher risk of hepatic steatosis compared to a non-obese individual. This important finding has a clinical implication in that, if an individual with the above mentioned variants can be identified at an early age, the significant modifying risk factors like higher waist circumference and triglyceride levels can be managed, thus reducing the predisposing risk component because of the variants in SNPs and thereby delaying the onset of hepatic steatosis.
Haplotype data for the three SNPs on PNPLA3, SAMM50 and PARVB suggests that three SNPs might be linked as heterozygous or mutant variant carriers were overrepresented in the NAFLD group (63 counts) compared to the control group (8 counts) (Table 5).
A recent study[29] from North India reported a higher frequency of CG and GG genotypes of rs738409 polymorphism in the PNPLA3 gene in North Indians and a significant association of the genotype to ALT (P = 0.003) and AST levels (P = 0.04). The values of triglycerides were slightly higher in the cases but were not significantly different in comparison to controls. This study is in agreement with the above study from the North Indian center with respect to the PNPLA3 polymorphism and its association with NAFLD and ALT levels. Our study also found a significant association of higher triglyceride levels with rs738409 polymorphism. However, the present study has looked at an additional 18 polymorphisms, which is by far the most comprehensive pooled genetic analysis taken up in Indian subjects with NAFLD.
To estimate the strength of the relationship between several independent variables and a continuous dependent variable, multiple logistic regression analysis was done with significant SNPs and patient characteristics like triglyceride levels, BMI from the univariate analysis as the independent variables and NAFLD as the dependent variable. While high levels of BMI, triglyceride levels, waist circumference both in males and females, ALT levels and variant SNPs in PARVB, SAMM50, NCAN, intronic SNP rs2499604, PZP and PNPLA3 were significantly associated with NAFLD when univariate analysis was done, only variants in PZP and PNPLA3 genes and high triglyceride levels were significantly associated with NAFLD when multivariate analysis was done, suggesting that these three are independent risk factors to predict hepatic steatosis and that the others probably interact with the modifying risk factors like BMI and waist circumference in the causation of NAFLD.
In conclusion, an analysis between the control and NAFLD groups revealed significant differences in BMI, triglyceride levels, waist circumference in both males and females and ALT levels with higher levels associated with the NAFLD group. Variant SNPs in NCAN and PNPLA3 genes were significantly associated with high ALT levels, which are the clinical phenotype of hepatic necroinflammation state, and SNPs in APOC3, PNPLA3, EFCAB4B and COL13A1 were associated with higher triglyceride levels.
The authors acknowledge the individuals that consented to participate in the study. They acknowledge Dr. HVV Murthy, Statistician, Asian Healthcare Foundation, for help with the statistical analyses and Dr. Rupjyothi Talukdar for his critical and constructive comments on the manuscript.
Non-alcoholic fatty liver disease (NAFLD) encompasses a spectrum of conditions associated with lipid deposition in the hepatocytes, ranging from simple steatosis (fatty liver) to non-alcoholic steatohepatitis (fatty changes with inflammation and hepatocellular injury or fibrosis), to advanced fibrosis and cirrhosis. It is the most common cause of liver disease, with a prevalence of 25%-30% in the general population. The presence of metabolic syndrome is the most common risk factor for NAFLD and it is now believed that NAFLD is the hepatic manifestation of metabolic syndrome. The other important risk factors are obesity, type-2 diabetes, total parenteral nutrition, jejunoileal bypass operation and use of certain medications. However, genetics play an important role in NAFLD and it is believed that 26%-35% of the patients who develop NAFLD have an underlying genetic component. So, it is important to identify the genetic aspects of the disease and their environmental interactions for better management of the disease.
Studies have identified that variant single nucleotide polymorphisms (SNPs) in genes, namely PNPLA3, NCAN, glucokinase regulatory protein, lysophospholipase-like protein 1, FDFT1, COL13A1, SAMM50, PARVB and pregnancy zone protein (PZP), were associated with NAFLD. A pooled genetic study was carried out by identifying significant SNPs from genome wide association studies and this study identified SNPs which are associated with Indian NAFLD. Apart from these associations, variant SNPs which contribute to hypertriglyceridemia, and alanine transaminase levels were also identified. By genotyping for these SNPs, an individual’s predisposing risk can be identified at an early age and lifestyle-based modifications would ensure delayed onset of fatty infiltration.
Susceptibility loci for Indian NAFLD have been identified for the first time. The genotype data can be used in early identification and better management of the disease.
Genome wide association study is the examination of many common genetic variants known as SNPs (single nucleotide polymorphisms) in two sets of individuals. One set of individuals with disease and the other set without the disease are compared for a large number of SNPs (approximately 9 lakhs) and analysis is done to identify those SNPs with a higher frequency in the disease group and these SNPs are said to be associated with the disease.
This well written and interesting pilot study of genetic susceptibility of NAFLD in an Indian population has shown that multiple SNPs and loci are involved in the development of NAFLD. Variant SNPs in PZP and PNPLA3 genes were found to be independent risk factors for the development of NAFLD. PARVB, SAMM50, neurocan and intronic SNP rs2499604 were significant risk factors along with other associations. So, genetics play an important role along with metabolic factors in the development of NAFLD. These findings may add a new level to the existing knowledge about the genetic basis of NAFLD, especially in the Indian population, and be valuable for clinical interference.
P- Reviewers: Ahmed M, Chang CJ, Fan JG, Mascitelli L, Milic S S- Editor: Wen LL L- Editor: Roemmele A E- Editor: Liu SQ
| 1. | Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592-1609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1226] [Cited by in F6Publishing: 1253] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 2. | Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585-1592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 319] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 3. | Wagenknecht LE, Palmer ND, Bowden DW, Rotter JI, Norris JM, Ziegler J, Chen YD, Haffner S, Scherzinger A, Langefeld CD. Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: the Insulin Resistance Atherosclerosis Family Study. Liver Int. 2011;31:412-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904-912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 278] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 5. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2233] [Cited by in F6Publishing: 2314] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 6. | Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 704] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 7. | Kawaguchi T, Sumida Y, Umemura A, Matsuo K, Takahashi M, Takamura T, Yasui K, Saibara T, Hashimoto E, Kawanaka M. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS One. 2012;7:e38322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 8. | Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, Taylor KD, Wilson L, Cummings OW. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 2010;139:1567-1576, 1576.e1-e6. [PubMed] [Cited in This Article: ] |
| 9. | Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, Dziura J, Lifton RP, Shulman GI. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082-1089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 318] [Cited by in F6Publishing: 330] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 10. | Agarwal AK, Jain V, Singla S, Baruah BP, Arya V, Yadav R, Singh VP. Prevalence of non-alcoholic fatty liver disease and its correlation with coronary risk factors in patients with type 2 diabetes. J Assoc Physicians India. 2011;59:351-354. [PubMed] [Cited in This Article: ] |
| 11. | Sanyal AJ. AGA technical review on nonalcoholic fatty liver disease. Gastroenterology. 2002;123:1705-1725. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 759] [Cited by in F6Publishing: 752] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 12. | Singh SP, Nayak S, Swain M, Rout N, Mallik RN, Agrawal O, Meher C, Rao M. Prevalence of nonalcoholic fatty liver disease in coastal eastern India: a preliminary ultrasonographic survey. Trop Gastroenterol. 2004;25:76-79. [PubMed] [Cited in This Article: ] |
| 13. | Amarapurkar D, Kamani P, Patel N, Gupte P, Kumar P, Agal S, Baijal R, Lala S, Chaudhary D, Deshpande A. Prevalence of non-alcoholic fatty liver disease: population based study. Ann Hepatol. 2007;6:161-163. [PubMed] [Cited in This Article: ] |
| 14. | Uchil D, Pipalia D, Chawla M, Patel R, Maniar S, Narayani A. Non-alcoholic fatty liver disease (NAFLD)--the hepatic component of metabolic syndrome. J Assoc Physicians India. 2009;57:201-204. [PubMed] [Cited in This Article: ] |
| 15. | Duseja A. Nonalcoholic fatty liver disease in India - a lot done, yet more required! Indian J Gastroenterol. 2010;29:217-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Mohan V, Farooq S, Deepa M, Ravikumar R, Pitchumoni CS. Prevalence of non-alcoholic fatty liver disease in urban south Indians in relation to different grades of glucose intolerance and metabolic syndrome. Diabetes Res Clin Pract. 2009;84:84-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 17. | Kumar R, Rastogi A, Sharma MK, Bhatia V, Garg H, Bihari C, Sarin SK. Clinicopathological characteristics and metabolic profiles of non-alcoholic fatty liver disease in Indian patients with normal body mass index: Do they differ from obese or overweight non-alcoholic fatty liver disease. Indian J Endocrinol Metab. 2013;17:665-671. [PubMed] [Cited in This Article: ] |
| 18. | Hepatic steatosis Ultrasound Images assessment procedures manual. Available from: http: //www.cdc.gov/nchs/data/nhanes/nhanes3/Hepatic_steatosis_Ultrasound_Procedures_Manual.pdf. [Cited in This Article: ] |
| 19. | Hernaez R, Lazo M, Bonekamp S, Kamel I, Brancati FL, Guallar E, Clark JM. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology. 2011;54:1082-1090. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 864] [Cited by in F6Publishing: 975] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 20. | WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7065] [Cited by in F6Publishing: 7538] [Article Influence: 376.9] [Reference Citation Analysis (0)] |
| 21. | Waist Circumference and Waist-Hip Ratio. 2008;8-11. [Cited in This Article: ] |
| 22. | Kang HS, Um SH, Seo YS, An H, Lee KG, Hyun JJ, Kim ES, Park SC, Keum B, Kim JH. Healthy range for serum ALT and the clinical significance of “unhealthy” normal ALT levels in the Korean population. J Gastroenterol Hepatol. 2011;26:292-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289-300. [Cited in This Article: ] |
| 24. | Eliades NG, Eliades DG. Haplotype analysis: software for analysis of haplotypes data. Distributed by the authors. 2009;. [Cited in This Article: ] |
| 25. | Ling TY, Huang YH, Lai MC, Huang SS, Huang JS. Fatty acids modulate transforming growth factor-beta activity and plasma clearance. FASEB J. 2003;17:1559-1561. [PubMed] [Cited in This Article: ] |
| 26. | Olski TM, Noegel AA, Korenbaum E. Parvin, a 42 kDa focal adhesion protein, related to the alpha-actinin superfamily. J Cell Sci. 2001;114:525-538. [PubMed] [Cited in This Article: ] |
| 27. | Humphries AD, Streimann IC, Stojanovski D, Johnston AJ, Yano M, Hoogenraad NJ, Ryan MT. Dissection of the mitochondrial import and assembly pathway for human Tom40. J Biol Chem. 2005;280:11535-11543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 141] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 28. | Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, Teranishi H, Mizusawa S, Ueno T, Chayama K. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783-792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Bhatt SP, Nigam P, Misra A, Guleria R, Pandey RM, Pasha MA. Genetic variation in the patatin-like phospholipase domain-containing protein-3 (PNPLA-3) gene in Asian Indians with nonalcoholic fatty liver disease. Metab Syndr Relat Disord. 2013;11:329-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |