Copyright
©The Author(s) 2025.
World J Gastroenterol. Jul 14, 2025; 31(26): 105656
Published online Jul 14, 2025. doi: 10.3748/wjg.v31.i26.105656
Published online Jul 14, 2025. doi: 10.3748/wjg.v31.i26.105656
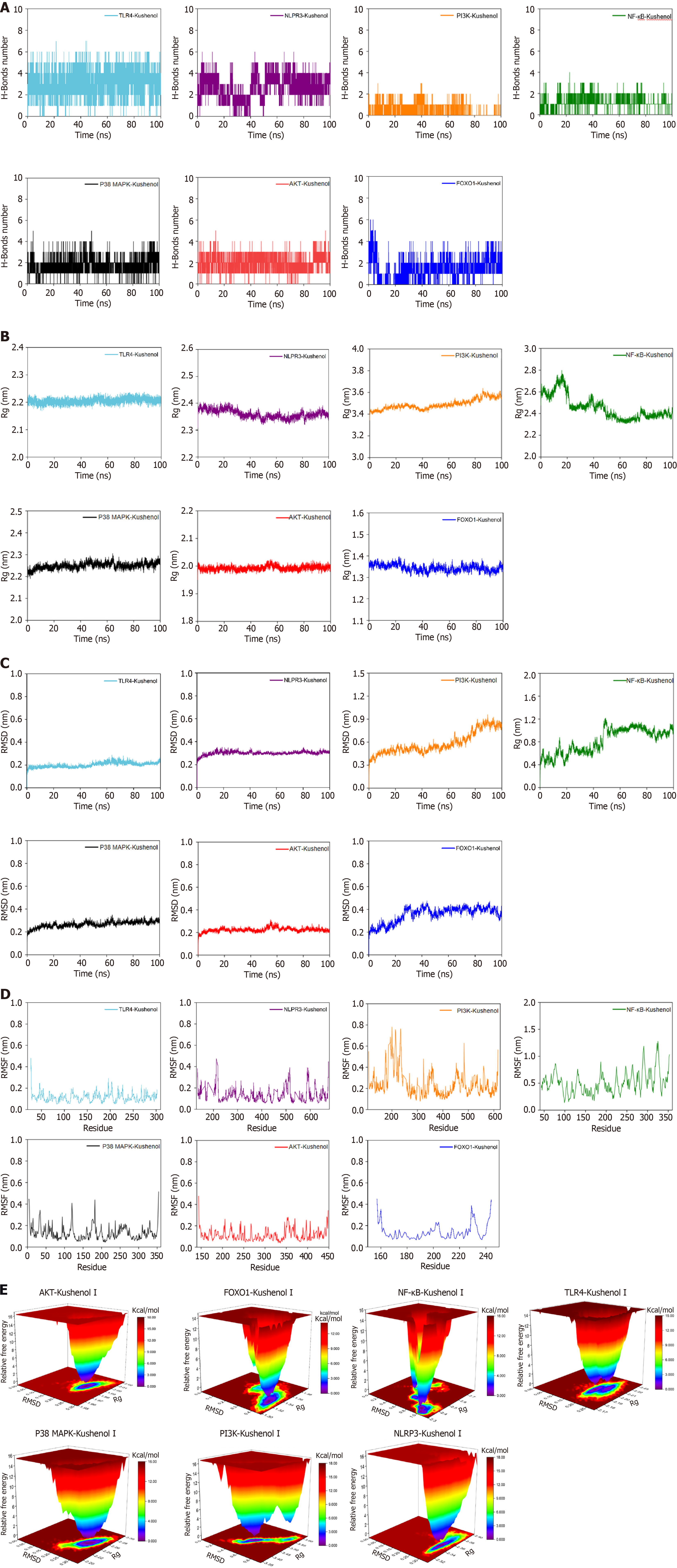
Figure 3 Molecular dynamics simulation results of kushenol I interacting with protein kinase B, forkhead box O1, nuclear factor kappa B, NOD-like receptor thermal protein domain associated protein 3, phosphoinositide 3-kinase, p38 mitogen-activated protein kinase, and Toll-like receptor 4.
A: Fluctuation curves of hydrogen bond numbers; B: Radius of gyration curves; C: Root mean square deviation curves; D: Root mean square fluctuation curves; E: Free energy landscape. AKT: Protein kinase B; FOXO1: Forkhead box O1; NF-κB: Nuclear factor kappa B; NLRP3: NOD-like receptor thermal protein domain associated protein 3; p38 MAPK: p38 mitogen-activated protein kinase; PI3K: Phosphoinositide 3-kinase; Rg: Gyration radius; RMSD: Root mean square deviation; RMSF: Root mean square deviation; TLR4: Toll-like receptor 4.
- Citation: He XD, Li M, Zuo XD, Ni HY, Han YX, Hu YK, Yu J, Yang XX. Kushenol I combats ulcerative colitis via intestinal barrier preservation and gut microbiota optimization. World J Gastroenterol 2025; 31(26): 105656
- URL: https://www.wjgnet.com/1007-9327/full/v31/i26/105656.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i26.105656