Published online Jun 7, 2021. doi: 10.3748/wjg.v27.i21.2681
Peer-review started: January 13, 2021
First decision: March 29, 2021
Revised: March 29, 2021
Accepted: April 21, 2021
Article in press: April 21, 2021
Published online: June 7, 2021
Artificial neural networks (ANNs) are one of the primary types of artificial intelligence and have been rapidly developed and used in many fields. In recent years, there has been a sharp increase in research concerning ANNs in gastrointestinal (GI) diseases. This state-of-the-art technique exhibits excellent performance in diagnosis, prognostic prediction, and treatment. Competitions between ANNs and GI experts suggest that efficiency and accuracy might be compatible in virtue of technique advancements. However, the shortcomings of ANNs are not negligible and may induce alterations in many aspects of medical practice. In this review, we introduce basic knowledge about ANNs and summarize the current achievements of ANNs in GI diseases from the perspective of gastroenterologists. Existing limitations and future directions are also proposed to optimize ANN’s clinical potential. In consideration of barriers to interdisciplinary knowledge, sophisticated concepts are discussed using plain words and metaphors to make this review more easily understood by medical practitioners and the general public.
Core Tip: This review summarizes the current achievements and existing limitations of artificial neural networks (ANNs) used in gastrointestinal (GI) diseases. The future directions of ANNs are also discussed to provide references for promoting its clinical value. To make this review readable, we introduce the basic knowledge of ANN and illustrate the contents from the perspective of gastroenterologists. ANN is believed to play a critical role in clinical practice of GI diseases.
- Citation: Cao B, Zhang KC, Wei B, Chen L. Status quo and future prospects of artificial neural network from the perspective of gastroenterologists. World J Gastroenterol 2021; 27(21): 2681-2709
- URL: https://www.wjgnet.com/1007-9327/full/v27/i21/2681.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i21.2681
The concept of artificial intelligence (AI) was first proposed at Dartmouth Conferences in 1956. Although definitions of AI vary, it is universally accepted that AI is designed to perform tasks that were previously fulfilled only by human intelligence. The debate on future directions of AI continues today. In the past few decades, AI has only been used to construct complex mathematical models. With the concurrent advancement of hardware, data, operation, and algorithm, AI has exhibited incomparable significance in many fields, such as fingerprint identification, information retrieval, and language translation. From 2016 to 2017, Robot AlphaGo driven by AI continuously defeated two world class Go grandmasters, which improved the connatural cognizance towards computer functions and drove us to reconsider the relationship between AI and human beings in the future.
As a branch of computer science, the integration of the cross-curricular interests of AI provides a strong boost for medicine. Many studies have focused on AI-aided screening, prevention, diagnosis, treatment, and health management. Since the MYCIN system was invented for clinical decisions with regard to infectious diseases in the 1970s[1], multiple systems supported by AI have been produced to meet the needs of medical care. For example, the AlmeHealthCoach system can automatically arrange medication regimens for patients at home. MedicalGraph can analyze the medical records and relevant laboratory tests to predict patients’ disease risk. These productions also performed well in the field of medical robots, imaging diagnosis, and research on new drugs. Such progress has broken through unwavering predicaments of current medicine.
Artificial neural networks (ANNs) belong to a subtype of AI and have been used in many subspecialties of clinical medicine, including pathology[2], radiology[3], cardiovasology[4], neurology[5], orthopaedics[6], and gastroenterology[7]. Gastrointes
This review focuses on the application of ANNs in GI diseases. We first introduce the basic knowledge of ANNs to help readers with a foundation for learning. The current achievements of ANNs are systematically summarized based on their applications. The characteristics, limitations, and future directions of research are then proposed from the perspective of gastroenterologists.
Machine learning (ML) is a method that endows computers with the ability to actively analyze data. If AI is a metaphor for one person, ML is like the brain of that person. The birth of ML serves as the fruit of multidisciplinary crosstalk, including statistics, probability theory, approximation theory, and algorithm complexity. The intrinsic connection of variables can be deduced based on learning experiences, as with a reasoned detective to reconstruct the crime from known facts. The cultivation of a detective requires the accumulation of basic knowledge and experience in handling many experiences. The detective will be qualified to participate in cases only after he receives adequate training and passes exams. Similarly, ML algorithms receive many data with known answers, and the resulting operation formula can be constructed to manage specific problems. This process is defined as the training phase. Different from classical linear functions, ML is automatic and unobservable. After training, ML algorithms will be tested with external datasets during validation phase, from which the test results are critical indicators of model efficiency and accuracy. Common algorithms for ML include ANNs, decision trees, naïve Bayes, support vector machines (SVMs), random forest, and expectation maximization.
Based on the learning approaches, ML can be typically divided into four subfamilies: (1) Supervised; (2) Unsupervised; (3) Semi-supervised; and (4) Rein
Unsupervised ML refers to a learning dataset without effective labels. This learning approach is typically used for disorderly and unsystematic data. Clustering is the representative use of unsupervised ML and can help uncover innate models and groups based on exploratory data. Taking next-generation sequencing as an example, tens of thousands of gene expression levels are listed. Genes will be clustered based on expression similarities and the process does not require prelabelled data. Additionally, unsupervised ML can be used to group and label datasets for next-step supervised algorithms.
Semi-supervised learning serves as an important helper for pattern recognition. It combines supervised and unsupervised approaches that flexibly use labelled and unlabeled data, thus integrating the advantages of both types. During semi-supervised learning, unsupervised parts can save the cost of human labor for labelling or classifying mass data, as well as decrease the difficulties of data handling, while the supervised part can guarantee model accuracy. Self-training, transductive learning, and transductive SVM are all types of semi-supervised learning.
Reinforcement learning is copied from the psychological process of acquired behaviors. Like training pets, any preset data are not required. Algorithms are gradually fed by signals in each layer and then receive “rewards” or “penalties” instantaneously after evaluating errors between true values and output data. The efficiency of algorithms can be adjusted during learning, and the core rules of reinforcement learning is maximizing rewards and minimizing penalties. How to obtain optimal balances and reliability allocation remains the most challenging problem for investigators. Currently, computers can independently solve complex problems based on reinforcement learning, as in Robot AlphaGo.
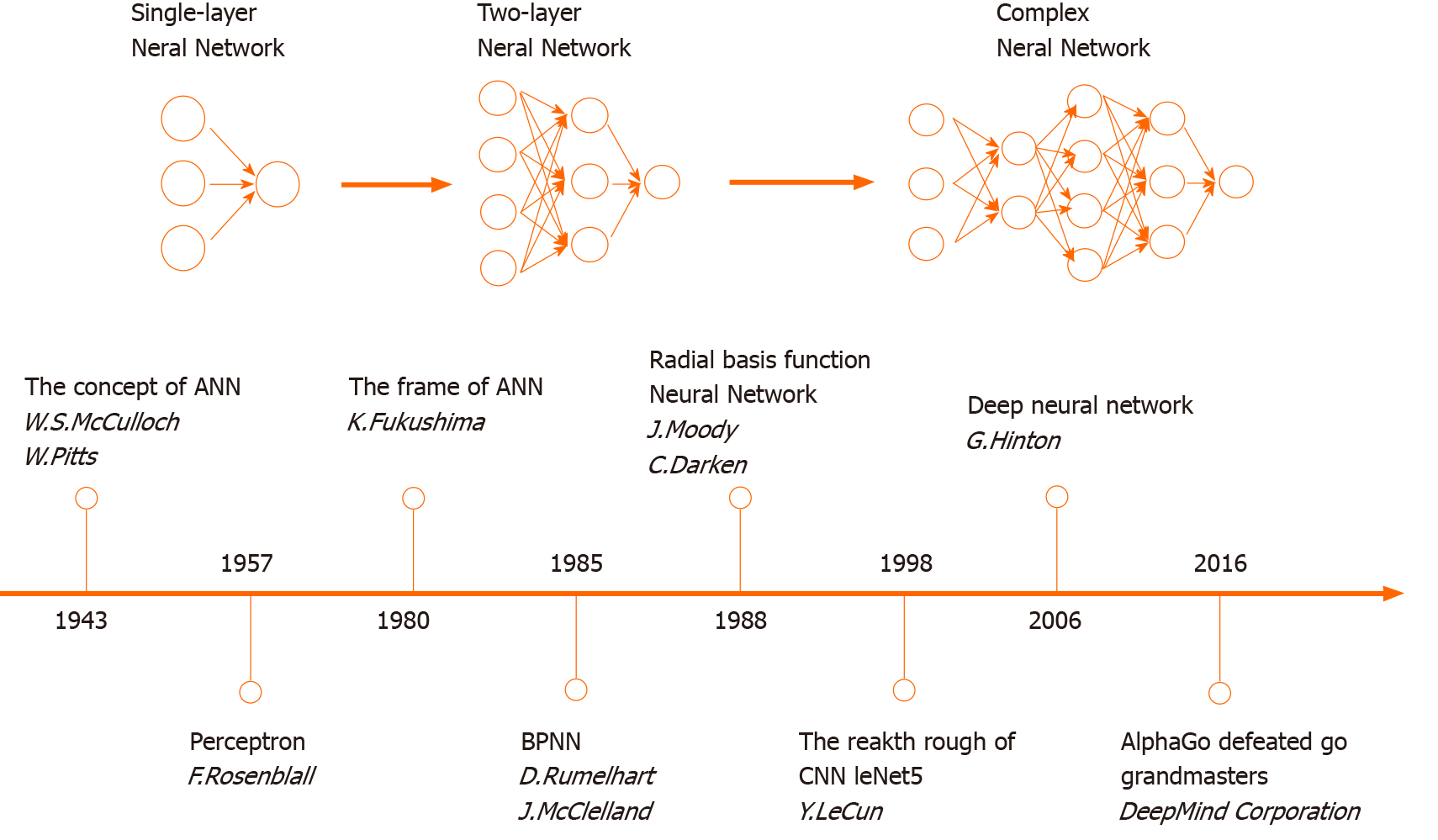
The concept of ANN was first proposed in 1943. The development of ANN models has complicatedly approached from then on. In the 1980s, the rapid progress of ANN algorithms boosted a modern revolution (Figure 1). The design of ANNs is based on the human brain’s neural network. An ANN is primarily composed of many neurons. The data flow (i.e., the signal) is passed and programmed across neural nodes. Neurons in the different layers have their own missions to solve problems, which can be analogous to factory production lines. The deconstruction of missions for data analysis provides the ability to find the optimal solution within the shortest time. As a type of parallel distributed system driven by mass data, ANNs are free from the requirements of logical or mathematical associations known beforehand.
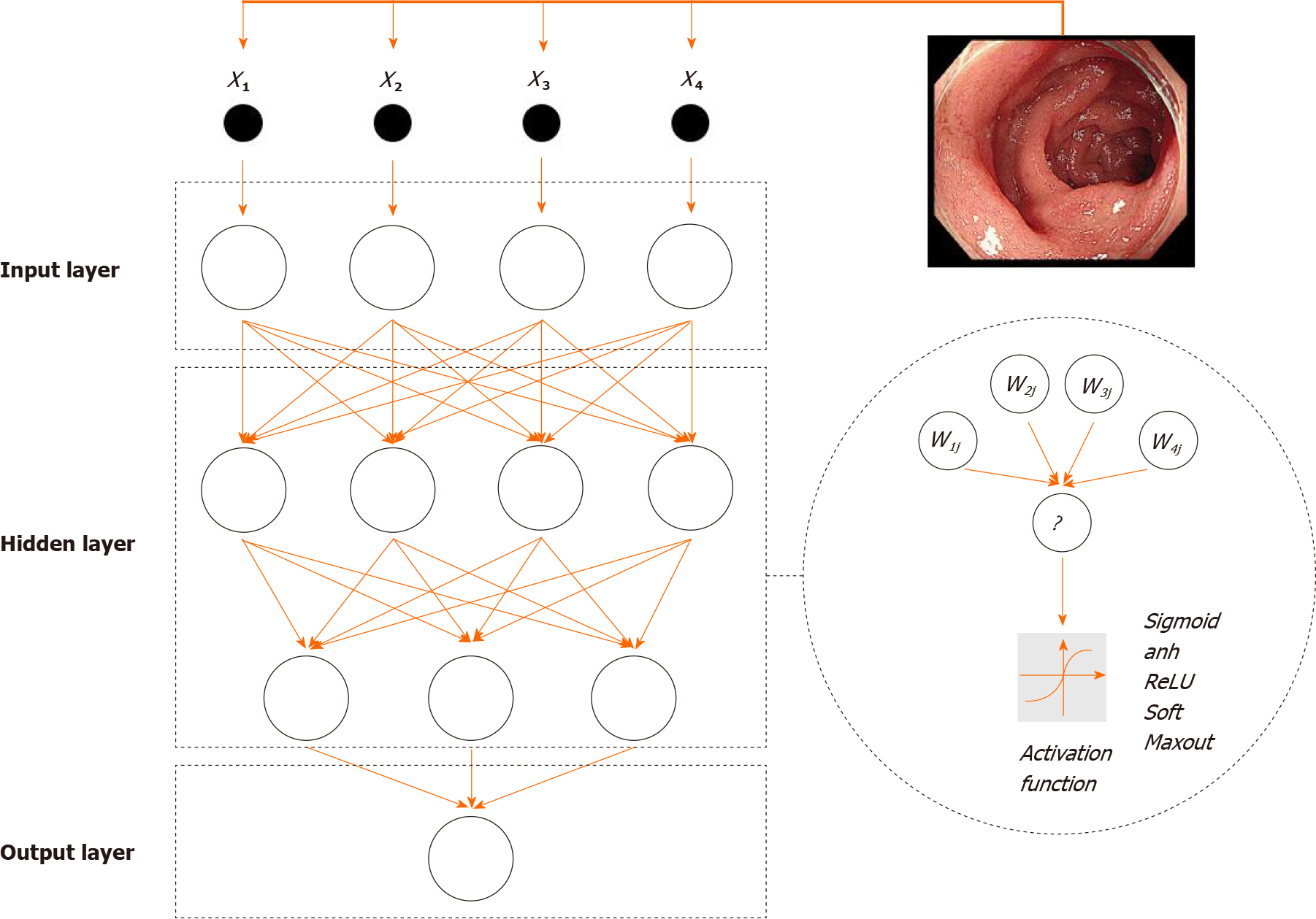
The structure of an ANN can be divided into three parts (Figure 2). First, input layers are responsible for receiving signals from specific datasets and serve as the “eye” of ANN to view the external environments. Second, hidden layers are predominant stages of ANNs and hinge on associations between input and output data. Raw data from input layers will be processed by neurons, and the numbers of hidden layers are always more than one. Missions for ANNs can be subtly disassembled and distributed into several layers. The multilayer structure promotes the efficiency of data analysis. Neurons in layers have serial connections based on settled sequences rather than parallel or parallel-series connections. Last, output layers make the final decisions and output results.
Common terms that describe shapes and capabilities of ANNs are listed (Table 1). The depth determines the efficiency of ANNs. The simplest ANN is a single-layer network (i.e., a perceptron neural network) and has one input layer, one output layer, and one hidden layer. A single-layer network can only be used to describe a single dichotomy based on a single factor, thus making most problems beyond its capabilities. Amplifying ANN depth can geometrically augment competence of fitting complicated features. Researchers began to create ANN models with multiple layers. Even in 2016, He et al[10] reported an ANN model with over 1000 layers[10]. However, excessive ANN depth may lead to system instability and loss of superficial learning abilities.
| Term of ANN | Specific meaning |
| Size | Number of neurons in the whole model |
| Width | Number of neurons in the one layer |
| Depth | Number of layers |
| Framework | Arrangement methods of layers and neurons |
| Capability | The reflection contents of reality by the specific model |
Each layer has a definite mission if the dataset volume and ANN depth are set. Width can be compared to the number of workers in a production line. Adequate workers will improve the qualities of tasks, while labor redundance will yield negative effects. A broad ANN width enhances the access to different data features, while a narrow width leads to low efficiency of feature extraction and excessive hardware burden. The concept of size describes the accumulation of both depth and width, which is an important index when evaluating an ANN’s capability.
Additionally, ANNs can be divided into two major categories based on the directions of signal flow between hidden layers. There are many differences between the two types of ANNs (Table 2). In the feedforward neural network (FNN), data flow is unidirectional from the upper levels to the lower levels without feedback; no connections exist between neurons in any given layer. In terms of system learning, FNN is convenient for reprogramming and can process non-linear questions. The other one is feedback neural network. One neuron in feedback neural network can pass its output data to the other neurons in the same or upper layers. The algorithms will be simultaneously adjusted by signals from other neurons based on a pre-knowledge dataset. Repeated calibration contributes to the excellent robustness and accuracy of the ANN. A feedback neural network is typically used for image analysis, diagnosis, and outcome prediction.
| Category | Feedforward neural network | Feedback neural network |
| Signal direction | Unidirectional | Unidirectional/bidirectional |
| Operation time | Short | Long |
| Feedback by output signal | No | Yes |
| Structural complexity | Simple | Complicated |
| Memory time | Short-term or none | Long-term |
| Applied ranges in medicine | Wide | Limited |
| Application | Perceptron network, back propagation network, radial basis function network | Recurrent neural network, Hopfieid network, Boltzmann machine |
Approaches of data processing in neurons can be defined as activation functions, and are like “all-or-none” principles of brain neurons. Activation functions primarily include sigmoid, relu, and tanh operations. Linear functions are excluded from activation functions because multiple overlays of linear functions can be compressed into one function. Accumulating studies have demonstrated that the nonlinear mapping ability of ANNs compensates for the boundedness of linear discriminant analysis. Otherwise, the “one-to-many” relationship between neurons guarantees the learning efficiency of the ANN. One neuron layer can simultaneously pass signals to more than one neuron. Importantly, signal flow is unequally allocated to each neuron in the next layer. The flow inequality makes the ANN adjustable to external environments. Before the computation of activation functions, data will be adjusted according to weight factors, each of which has a corresponding relationship with a specific neuron. Weight factors serve as the controllers of data flow. Larger weight factors have a greater influence on the output results. Conversely, setting a weight factor equal to 0 can abrogate the functions of the corresponding neuron.
There are a wide range of ANN categories used in basic and clinical medicine, and each contains a specific algorithm that has distinct advantages over the others. Comprehension and discrimination of ANN categories can provide a good command of research frontiers, and ensure that ANNs are used in the correct situations. Brief introductions of common ANN methods are as follows.
Convolutional neural network: Convolutional neural networks (CNNs) are the most popular type of neural network nowadays due to their additional convolutional layers and pooling layers. Convolution and pooling are complex concepts in computer science. Briefly, convolutional layers can extract features from a dataset because neurons in this layer are responsible for collecting and analyzing information located in specific areas. These neurons can be analogue to cells in the visual cortex of a human brain and act as windows that look out across the external environment. Data is then sent to the pooling layers after convolution, where the features of the dataset are filtered and compressed. Pooling layers reduce background and texture information at the cost of partial data loss. Then, the data will be processed in classic neural networks.
The number of CNN layers is relatively flexible. The accuracy and performance, however, are limited in CNNs with shallow structures. The majority of CNNs belong to deep CNNs. CNNs are applicable in numerous fields, such as image classification, objective recognition, and natural language processing. These applications require learning features from mass data and being able to generalize results. Also, fine-grained recognition of medical images is the core difficulty of CNNs for feature extraction and can be finished using handcrafted or unsupervised CNNs. The extraction efficiency of unsupervised CNNs outperforms handcrafted CNNs[11].
BPNN: BPNN is a type of multi-layer FNN. BPNN algorithms can be divided into two stages: Forward propagation and back propagation. For forward propagation, signals are handled from input layers to hidden layers and then to output layers. If errors between the real output and expected output are not acceptable, the error values will undergo back propagation, where the error values will be reversely transmitted layer by layer and then equally split into all involved neurons. Neurons automatically use revisions based on feedback signals; thus, BPNN can learn and apply mapping relations without clarifying mathematical derivation beforehand. The simplicity and accuracy of BPNNs have allowed them to be widely used in medicine, particularly for the diagnosis and prediction of patient outcomes.
Bayesian neural network: The training phase of the ANN requires a certain amount of data, and small training datasets tend to impair the efficiency and practicability of most ANNs. However, sufficient sample size is not always available. Bayesian neural networks (BNNs) have been designed for such situations. BNNs are based on weight factors that describe probabilistic ranges that are subject to Gauss distribution instead of being constants. The parameters optimized by back propagation are the means and variances of the weight factors, and can increase the confidence of each item of data, increasing the dataset robustness. Recently, BNNs have exhibited great potential in analyzing image features and constructing disease models.
Recurrent neural network: Data are frequently independent in a dataset, and the input sequence makes no difference in neural networks. However, there are sometimes internally logical relations between input information. For example, a set of computed tomography (CT) images contain many layers, and the observation of images in order can dynamically describe the sizes and shapes of lesions. Thus, treating these images as independent data would lose much information. Recurrent neural network (RNN) was invented to overcome this limitation. As introduced before, the values in hidden layers are under the control of weight factors. In RNNs, weights are regulated by not only values of this time, both at the current time step and the last time step; thus, the output values of RNNs are affected by the input sequence of the dataset. RNNs have been widely used to manage datasets with successional subjects.
GI diseases primarily refer to acute and chronic disorders in the stomach and intestinal tract. The diagnosis and prevention of GI diseases remain challenging for clinical practice based on three aspects: Anatomy, symptoms, and pathogenesis. First, regarding anatomy, the digestive tract is exposed to the external environment all the time; however, most of the digestive tract is beyond our visual observation. Thus, the GI tract is vulnerable to various irritants, but pathologic deterioration cannot be easily detected. Second, the symptoms of GI diseases are atypical. Patients with early-stage GI tumors tend to underestimate their disease severity and miss the best opportunities for treatment. Concurrently, symptoms are not qualified as critical indicators of diagnosis and outcome. Clinicians must use imaging examinations to further clarify disease conditions. Imaging interpretation is somewhat subjective, relying on experiences of radiologists. The rates of missed and delayed diagnosis remain high. Last, in terms of pathogenesis, GI diseases are multifactor diseases including heredity, food, microbes, and neuropsychic factors. Single preventive methods are ineffective for the treatment of GI diseases. However, the application of AI has gradually broken through this dilemma.
Pathological diagnosis is the gold standard for GI diseases, which has significant implications for diagnosis, therapeutic directions, and outcome prediction. This work is highly dependent on the subjective judgement of pathologists. Accuracy is susceptible to recognitive capability, professional experience, and fatigue levels. The misdiagnosis of pathological images remains beyond clinical requirements. With the rapid development of digital pathological sections, the transformation from qualitative to quantitative analysis has become a research topic. ANN models are effective at computer-aided pathological diagnosis. Karakitsos et al[12] tried to explore the potential of ANNs to discriminate benign from malignant gastric cells, and their model accuracy reached 97%[12]. Despite concentrating on cellular morphology, the capability of interpreting pathological images has been revealed.
Haematoxylin-eosin (H&E) staining is the most common method for discriminating benign and malignant lesions. For GC, most studies used CNN models to construct diagnostic systems for H&E staining[13-17]. In 2017, Sharma et al[17] reported the first study about computer-aided classification in H&E staining of GC. Regretfully, their system exhibited an accuracy of only 69.9%, far from the clinical standard[17]. Rapid progress has been made during the past three years. A CNN model fed by over 2000 H&E slide images achieved a near 100% sensitivity and 80.6% specificity[13]. Such optimal sensitivity showed that this model has potential in preliminary screening of H&E staining. Kosaraju et al[14] developed a novel CNN model called Deep-Hipo, which could extract and capture features at both high and low magnification levels. The overall accuracy of Deep-Hipo was 93.7%[14]. Deep learning model was also constructed for the prediction of microsatellite instability in colorectal cancer. The model significantly outperformed pathologists[18]. Importantly, models detecting multiscale receptive fields can be extended for survival prediction, subtype classification of GC, and selection of sensitive drugs.
Most studies concerning colorectal cancer (CRC) have also focused on distinguishing between benign and malignant tissues. Based on existing results, the accuracies or area under the curve (AUC) values of ANN-based models all exceed 80%[19-25]. Colon glands are important structures and indicators for pathological assessment[26]. However, the variation of glands in slides has been a barrier both for pathologists and algorithms. CNN models invented for it got excellent results about gland segmentation and classification[27-30]. Sirinukunwattana et al[31] showed a CNN model that automatically detects cell nuclei in CRC. The combination of spatially constrained CNN and softmax CNN might become a useful tool for exploring the tumor microenvironment[31]. Interestingly, a study investigated the correlation between genomic and epigenetic alterations in CRC. CNN algorithms were used to predict RNA expression classifiers from H&E images. This work lays the foundation for a comprehensive integration of morphology and molecular features[32]. Immunohistochemical staining and fluorescence microscopy belong to members of pathological examinations, and ANNs distinctly improved their diagnostic perfor
In addition to the auxiliary role of ANNs in pathological images, Wei et al[19] compared the accuracy of their model with that of pathologists and there was no significant difference between them[19]. The process from image input to result output could be finished within 2 s[20,36,37], and the time for ANN handling single image is evidently shorter than that for pathologists. ANNs can also complete repetitive processes with high-quality results. Based on the report of Yoon et al[20], the training time was only approximately 10 d[20], while the training time of a skilled pathologist was based on the order of years. Thus, all evidence indicates the great potential of ANNs in fast pathological diagnosis.
X-ray, CT, and magnetic resonance imaging (MRI) are common radiological examinations. CT is accepted as first choice for GI diseases due to its high image quality, convenience, and price[38,39]. The increasing clinical demand for examinations places great burdens on radiologists. Experience-based diagnosis has innate limitations in accuracy and efficiency. This dilemma requires novel computer-aided models with the support of new techniques. Compelling evidence indicates that ANNs can provide outstanding values in auxiliary diagnosis[40,41], and are not limited to this field.
At the end of the last century, an Austrian group investigated the potential of ANNs in single-photon emission CT[42]. Despite the lack of further validation, this innovative research launched a sharp increase in related studies. Many of them aimed to detect primary lesions in CT images[43-47]. For example, a CNN-based model was trained with 288 CT images for polyp detection and achieved a sensitivity of 97%[47]. Studies published in earlier years used multiple types of ANNs, including BPNNs, massive-training ANNs, and BNNs[45,48-50]. In recent years, investigators have collectively turned to using CNNs[43,51,52]. This tacit alteration suggests that CNNs have fewer complications in automatic CT interpretations. The combination of ANNs and CT has also been used in the diagnosis of cancer metastasis and invasion depth[53-56], which are also important indicators of patient prognosis. A study even reported that CNN-based systems could distinguish benign and malignant lesions by CT[57]. More possible development directions of CT were discussed. In brief, the diagnostic value of CT has been tremendously strengthened with the support of ANNs.
MRI has a superior definition of abdominal viscera and presents more information to radiologists than CT. Most studies focused on the promotion of MRI in CRC. A CNN-based system created by Trebeschi et al[58] reached up to an AUC of 99% for advanced rectal cancer[58], and other recent studies all performed well in this field[59-61]. Certain studies have tried to explore new applications of MRI. CNNs could increase the diagnostic capabilities of 3D reconstruction[62]. Faster Region-based CNN was trained with 240 cases for evaluation of circumferential resection margins and achieved a 95.6% specificity[63]. Chemosensitivity prediction by MRI was also achieved with the support of algorithms[64,65]. KRAS mutation is an important biomarker of CRC targeted therapy[66,67]. Genetic testing is the gold standard of detecting KRAS mutation. Two Chinese groups demonstrated that deep radiomics signature serves as a promising tool for the prediction of KRAS mutations[68,69]. The progression of algorithms could also elevate the sensitivities of 3D Stomach MRI[70]. These achievements suggested the feasibility of non-invasive detection for medication guidance of GI tumors. Otherwise, the importance of ANNs in inflammatory bowel disease (IBD) diagnosis has been shown[71].
Endoscopy is universally accepted as a revolutionary application in gastroenterology. Due to the unique advantages of the GI tract, endoscopists can visually observe diseased regions, obtain biopsies, and excise small lesions. The extensive application of endoscopy has effectively reduced mortality and economic burdens worldwide[72,73]. However, artificial discrimination and classification lead to a relatively high incidence of misdiagnosis. Conversely, endoscopy is widely used to screen and exclude diagnosis for health checks and suspected cases. Endoscopic sensitivity, therefore, must be improved. Thus, numerous studies have focused on the application of ANNs in endoscopy, and occupy the largest proportion of existing research.
The detection of GI tumors has become the mainstream direction of endoscopy. Many studies have developed endoscopic ANN-based systems. Like other methods of image analysis, investigators prefer CNNs to construct computer-aided models when managing endoscopic results[74-78]. For example, Hirasawa et al[74] trained a CNN model with 13584 endoscopic images and effectively captured GC lesions with a diameter of > 6 mm as well as all invasive GC[74]. Bagheri et al[79] proposed a CNN-based system to analyze the color levels during colonoscopies. The stratification of color levels significantly promoted diagnostic values, achieving a 93% precision and an 82% Dice score[79]. Otherwise, the fast speed is another advantage of ANN. Models can analyze thousands of images in a short time[74,80-83], which benefits from retrospective checks of images and facilitates real-time endoscopic diagnosis. For example, Zhang et al[80] developed a CNN model for the detection of gastric precancerous diseases in 2017[80]. After 2 years, they developed a novel CNN-based system used for gastric polyps. This model achieved real-time analysis with 50 frames per second without a decline in diagnostic precision[84]. Intraoperative guidance and correction are urgently required. A deep CNN trained by 704 cases of colorectal adenomas, called ENDOANGEL, was published to help endoscopists monitor detection operations and avoid blind spots in real time[85]. This study is significant for computer-aided refinement of endoscopy.
Early screening has received much attention as people gradually attach importance to routine examinations. The atypical textures of lesions at an early stage pose a challenge for endoscopists. Numerous studies have aimed to improve the detection of GI polyps, precancerous lesions, and tumors at early stage using ANNs[9,78,86-92]. Interestingly, most research groups are from China and Japan, likely due to the regional characteristics of GI tumor incidence. The best performance of early detection was reported by Gao et al[78]. Their CNN system reached an overall accuracy of 98.7% and a specificity of 100%. ResNet50 stood out of five CNN models and performed the best in detecting CRC precancerous lesions with an accuracy of 93%[78]. Moreover, ANNs could assist in diagnosing Helicobacter pylori infection using the combination of blue laser imaging and linked color imaging[93-95]. This breakthrough undoubtedly expands the application ranges of endoscopy.
ANN application is not confined to traditional white-light endoscopy. Narrow-band imaging (NBI) is a new type of technique that can filter out broad-band spectrum in red, green, and blue colors, respectively. The concentrated layers of the GI mucosa can be altered by switching filtered wavelengths, endowing NBI endoscopy with unique strengths in detecting minor cancerous and even precancerous lesions. The integration of NBI and ANN has made amazing progress in early screening. Three studies respectively reported their models for the diagnosis of early GC[81,86,87]. For instance, Horiuchi et al[87] trained a CNN system that could differentiate between early GC and gastritis. This model achieved a high sensitivity but low specificity, yielding undesired accuracy[87]. In consideration of these fields of study, bias towards sensitivity is accepted. Trails concerning NBI colonoscopy were also performed and exhibited expectable results[7,96-98].
In recent years, wireless capsule endoscopy (WCE) has been developed for patient acceptance and the extension of detection zones. The small bowel, which is known as the blind spot of classical endoscopy, can be virtually examined only by WCE[99,100]. Thus, investigators use ANNs to enhance the capability of WCE diagnosis. A decade ago, Barbosa et al[101,102] began to develop an automatic detection system using a multilayer perceptron neural network[101,102]. Despite the limited size of the training dataset, this study proposed a novel direction for forthcoming models with great performance. With the development of WCE and algorithms, CNNs were shown to have sufficient potential to detect lesions in small-bowel WCE[8,83]. Particularly, Ding et al[8] trained a model with a large-sample dataset, achieving a sensitivity of 99.88%[8]. These achievements have changed the belief that the small bowel is the dead zone of GI tract examinations. However, two shortcomings of WCE still exist. First, WCE goes through a natural orifice, and its photograph angles are affected by GI peristalsis, body position, and contents within GI tract, presenting the possibility of losing sights. Second, during WCE movement, secreted mucus and chyme are likely to stick to WCE, decreasing image quality. ANNs may serve as an effective approach to solve these problems[103].
In addition to GI tumors, ANNs play an important role in the diagnosis of other acute and chronic GI diseases. Many studies concerning GI bleeding focused on WCE. The imaging features of serious GI bleeding are relatively distinct, while small lesions with bleeding are likely to be missed, particularly in WCE. The authors used CNN, BPNN, and probabilistic neural network (PNN) to optimize the WCE capabilities of instant diagnosis, which covered the stomach, small bowel, and colorectum. These ANN models were all reported to perform well[104-109]. Different from GI bleeding, progress on using ANNs to diagnose chronic atrophic gastritis is slow, likely because of atypical features and low rates of early screening. Two recent studies indicated the tremendous potential of CNNs in this field and showed that CNNs outperformed the expert endoscopists in identifying GI bleeding[110,111]. IBD is another indication of colonoscopy, and ANN was also used to improve the diagnostic value of endo
A research group conducted a retrospective study of 521 GC patients in 2005. Three mining techniques, including ANN, decision tree, and Logistic regression, were used to analyze clinical information and predict postoperative complications. ANN outperformed the other two methods[114]. Since then, many studies have focused on the clinical potential of ANNs in risk prediction. In particular, a large number of relevant reports have emerged in recent years, which is reflected in the rapid improvement of ML algorithms. ANN models for predicting cancer risks commonly include dozens of categorical variables, such as age, sex, oncology indicators, radiological tests, and therapeutic regimens[115-118]. Two Japanese groups combined clinical indicators and endoscopic images using CNNs to predict GC risks[94,119]. Genetic and microbial factors function as important regulators in carcinogenesis. Genomics and microbiome can exhibit the deep roots of GI tumors. Dadkhah et al[120] trained ANN models with the gut microbiome of 218 subjects to identify risks for colorectal polyps. Classification accuracy exceeded 75% based on home stool samples[120], which is expected as a novel method for convenient screening of colorectal polyps. mRNA profiles also have referential value in predicting the odds of malignant transformation[121,122]. The fruits of high-throughput screening techniques combined with deep analysis can depict the landscape of carcinogenesis. Subsequent etiology-based tests will contribute to revolutionary progress in precise prediction.
Metastasis is a critical index of therapeutic regimens and long-term prognosis. CT and MRI have detective limitations and only positive lymph nodes with swelling shapes can be observed, as well as distantly metastatic lesions. The results, however, always fall behind the real condition, misguiding surgical strategies and reducing survival benefits. Investigators have tried to use the advantages of ANNs to elevate the accuracy of the metastatic conditions in GI cancer. A German group used an FNN to predict lymph node metastasis of GC early in 2005[123], but the performance is poor, partly due to unadvanced algorithms and computers. With the development of ANNs, models included larger amount and more subtypes of clinical indicators. Their predictive efficiency has been augmented to reach clinical standards[124-127]. Two studies investigated ANNs used in identifying metastatic risks of CRC patients at T1 stage[128]. Although cases with metastatic T1 tumors are rare, the underlying mechanisms may be uncovered with the support of ANN. Kurokawa et al[128] first used BNN to construct a predictive model that involved a novel molecular target from basic medicine and showed great potential value[128]. The growth of novel biomarkers has recently provided more choices for GI diagnosis and prediction; however, few studies have emphasized the integration between these newly found targets and classical tumor biomarkers. Future development requires more investigation of ANNs to maximize the significance of existing fruits.
Survival time and quality of life are important factors that both patients and doctors consider. No efficient approaches have been developed to date, which is typically explained by disease heterogeneity, subpopulation differences, and medical experiences. These problems are likely based on one reason: Deficiency of data integration. Investigators rest their hopes for an accurate diagnosis on the excellent capabilities of information screening and automatic decision making. They used ANNs to weight many kinds of factors to quantify the survival outcome of cancer, GI bleeding, and IBD patients[106,129-133]. For GI cancer, certain studies compared ANN with TNM stage and models constructed by other ML methods. ANN typically exhibited better performance than Logistic regression, TNM stage, and even clinicians[134-136]. Competitions between different types of ANNs were also performed and the results were not consistent[137-139].
In addition to classical types of clinical information, certain novel biomarkers combined with ANNs have potential value in the prediction of GI disease risks and prognosis. Altomare et al[140] created a panel composed of exhaled volatile organic compounds (VOCs) from CRC patients. A PNN was used to synthetically analyze the levels of exhaled VOCs. This model could effectively identify CRC patients and predict the survival benefits from curative CRC surgery[140,141], which was a quantum leap in non-invasive screening. Several immune markers, including CD8, CD20, and CD68, combined with the proliferation marker Ki-67 by CNN showed great prognostic value in GC[135]. DNA aneuploidy, tumor-stroma ratio, and RNA sequencing were also proven to have great value in clinical prediction that were evacuated by ANN[142-144]. Otherwise, the guiding capabilities of ANNs in the prediction of both inpatient and outpatient services have been gradually revealed[145-148]. With the deep crosstalk between ANNs and medical needs, clinical tasks that were previously regarded as impossible or difficult are becoming feasible and easy.
Clinical decision making plays a conclusive role in the process. Numerous clinicians and scientists have been striving to promote standardized, precise, and individualized treatment. As the primary adjuvant regimens of GI cancer, chemotherapy should consider many factors, such as patient tolerance, pathological sensitivity, concrete dosage, and regimens. Accumulating evidence has showed that ANNs could help overcome the therapeutic limitations caused by individual differences and mass data. Evaluation of chemotherapeutic sensitivity with ANN models is a hot topic. To establish reliable prediction systems for locally advanced and metastatic CRC, several ANN models have been used to integrate clinical indicators[149,150]. Their accuracies were significantly better than those of clinicians. Moreover, deep learning of radiological images exhibited its potential value in assessing chemosensiti
The quality control of surgical guidance and supervision is highly dependent on the skills of clinical teachers. It is difficult to cultivate doctors who receive normalized training and guarantee surgical processes free from interference of uncertainties. Kitaguchi et al[156,157] created a novel CNN-based deep learning approach fed by intraoperative videos. It could automatically recognize the surgical phase and action with high accuracies of 81% and 83.2%, respectively[156,157]. These achievements will initiate a novel field of ANN application. Clinicians, especially young surgeons, will reap benefits from these technological innovations. Regretfully, few studies have been reported due to ethic restriction and complicated surgical situations. More attention and investment should be paid for developing efficient guiding systems that precisely match skills and customs of surgeons.
Basic research is the frontier where ANNs have been widely used. Inherent logic and correlation between variables can be easily shown by ANNs, which may be too difficult for human intelligence. Many repetitive tasks can be finished within a short time. In the past 10 years, the rapid progress of ANNs has launched another wave of research (Table 3). Existing studies can be divided into three aspects: (1) Elucidation of mechanisms of GI carcinogenesis; (2) Guidance of drug selection; and (3) Integration of fundamental research data. First, high-throughput screening is the fundamental method for selecting critical molecules and pathways of GI cancer. High-throughput techniques will produce extremely large amounts of raw data. It is impossible to precisely choose potential targets only by manpower. Bidaut et al[158] began to use ANNs to characterize stomach stem cells based on microarray experiments[158]. This technique showed great value in the basic research of GI physiology. A protein-based ANN classifier was successfully constructed to identify the origins of tumor samples[159], which may promote the application of mass spectrometry in precise tumor diagnosis. Cell counting serves as a fundamental task in basic and clinical research. The low efficiency of classical manual detection cannot meet the requirement of many experiments. CNNs can significantly improve cell counting technology on their own merits[160]. A CNN-based cell counter has already been commercialized. Moreover, CNN showed great performance in discovering lncRNA-disease associations[161,162].
| Ref. | Disease | Type of data | ANN technique | Application direction | Outcome |
| Bao et al[167], 2020 | CRC | Microsatellite instability from TCGA database | Multi-layer perceptron network | Prognostic prediction | 100% accuracy |
| Coppedè et al[189], 2015 | CRC | DNA methylation | AutoCM | Identification of connections between DNA methylation and CRC | A strong connection between the low methylation levels ofthe five CRC genes |
| Liu et al[190], 2004 | CRC | Gene signature from GEDatasets | Multi-layer network | Identification of latent marker genes of CRC | 91.94% accuracy |
| Berishvili et al[164], 2019 | CRC | Approximately 4000 complexes for which the data on the target binding constants | CNN | Screening filter for compoundprioritization | 73% Spearman rank correlation coefficient |
| Bloom et al[159], 2007 | CRC and GC | MS | Multi-layer network | Differentiation between 6 common tumor types | 87% accuracy |
| Dadkhah et al[120], 2019 | colorectal polyp | Gut microbiome | ANN developed by Orange data mining tool | Early screening using collected stool | > 75% accuracy |
| Chang et al[166], 2011 | CRC | miRNA profile | Not mentioned | Exploration of association between specific miRNAs and clinicopathological features | 100% accuracy of miRNA panel |
| Chen et al[191], 2004 | CRC | MS of serum protein pattern | Multi-layer perceptron network | Differentiation between CRC and healthy control | 91% sensitivity; 93% specificity; 0.967 AUC |
| He et al[121], 2020 | CRC and gastroesophageal cancer | Gene signature from TCGA database | Multi-layer network | Differentiation between types of cancer | CRC: 98.06% sensitivity; 96.88% precision. Gastroesophageal cancer: 94.89% sensitivity; 96.33% precision |
| Hu et al[192], 2015 | CRC | Gene signature from database of NCBI NLM NIH | S-Kohonen neural network | Prediction of recurrence using gene expressions | 91% accuracy |
| Kurokawa et al[128], 2005 | CRC | Gene signature of nodal metastasis | BNN | Prediction of metastatic potential of CRC at stage I | 88.0% sensitivity; 86.6% specificity; 0.904 AUC |
| Liu et al[160], 2019 | Cancer cell | Synthetic microscopic images from two publicly datasets | CNN | Automated counting of cancer cells | - |
| Ronen et al[193], 2019 | CRC | Gene signature from TCGA database | BNN | Stratification of CRC subtypes | - |
| Bilsland et al[194], 2015 | CRC | A virtual library of compounds | Perceptron network | Screen of Benzimidazolone inhibitors for CRC treatment | CB-20903630 was selected out for further validation of CRC treatment |
| Maniruzzaman et al[195], 2019 | CRC | Gene signature from patients | Fuzzy neural network | CRC classification | 99.84% sensitivity; 99.75% specific; 99.81% accuracy; 0.9995 AUC |
| Inglese et al[196], 2017 | CRC | 3D MS | Deep neural network (unsupervised) | Identification of metabolic heterogeneity | Up to 0.6991 Pearson's correlation |
| Shi et al[197], 2020 | CRC with liver metastasis | CT | ANN | Prediction of KRAS, NRAS and BRAF status | 0.95 AUC |
| Jiang et al[198], 2020 | GC | Two drug datasets | deep neural network | Prediction of drug-disease associations | 17 kinds of drugs that were screened out by ANN had been confirmed as anti-tumor drugs |
| Bidaut et al[158], 2009 | Stomach stem cell | Stemness signature | Perceptron network | Characterization of stem cells | - |
| Jing et al[168], 2019 | Calibration of laboratory markers | CA-724 | Radial basis function neural network | The effects of geographic factors on CA-724 | CA724 reference values show spatial autocorrelation and regional variation |
| Xiao et al[122], 2018 | GC | RNA-seq | Probabilistic neural networks (semi- supervised) | Diagnosis of cancer | 96.23% accuracy; 99.08% precision |
| Hang et al[144], 2018 | GC | MSI | Multi-layer perceptron network | Prognostic prediction | 0.81 AUC |
| Xuan et al[161], 2019 | GC | LncRNA profile | CNN | Prediction of GC | 0.977 AUC |
| Joo et al[163], 2019 | GC | Potential drugs from databases | CNN | Exploration of new drugs targeting | ANN-based model accurately predicts drug responsiveness as models previously reported |
| Liu et al[165], 2010 | GC | MS from GC patients | Supervised neural network | Early screening | 100% sensitivity; 75% specificity |
| Que et al[199], 2019 | GC | MS from GC patients and clinicopathological parameters | Single-layer neural network | Prediction of long-term survival | 0.82 AUC |
| Li et al[200], 2021 | GC | Gene Expression Omnibus database | ANN | Differentiation between GC and healthy tissues | 0.946 AUC |
Development of new drugs is an important direction of research. Computer-aided target selection has gradually replaced inefficient artificial experiments. Joo et al[163] aimed to screen compounds targeting dysfunction proteins to find new drugs for CRC. Preliminary selection was performed using ANN, and then a benzimidazolone compound was selected out for further cell toxicity tests. This study provides a good example for subsequent studies that explore novel medications. A CNN-based model, DeepIC50, was trained with 27756 features at molecular levels. It was proven to effectively predict three drug responsiveness classes for GC patients[163]. Further validation is required to determine whether the applicable ranges of DeepIC50 can extend to other types of tumors. Furthermore, tankyrase inhibitors have been verified as potential anticancer drugs. To confirm the activity ranking of tankyrase inhibitors in CRC treatment, Berishvili et al[164] developed an affinity prediction model based on CNN. The Spearman correlation of the model achieved 0.73, which was superior to those of the other three virtual screening approaches[164].
With the development of molecular biology, many types of high-throughput screening techniques have been widely used, including RNA-seq, protein mass spectrometry, metabolomics, microbiome, and noncoding RNA chips. Diverse types of ANN were employed to evacuate efficient targets from sequenced data
Although most studies paid their attentions to ANNs in GI cancer, the functions of ANNs in noncancerous GI diseases were also revealed. IBD is a collective name, covering ulcerative colitis and Crohn’s disease. Recent studies manifested the potential value of ANNs applied in clinical practice of IBD (Table 4), which preliminarily focused on the following two parts: (1) Differential diagnosis. ANNs could help distinguish between IBD, irritable bowel syndrome, and healthy controls. The sensitivities reported by the studies were all over 75%[169-172]. The data suggested the translational potential in diagnostic assistance of ANNs; and (2) Prediction of therapeutic efficacy. Surgery, hormone therapy, immunosuppressors, and targeted drugs are alternative regimens for IBD therapy. ANN models were proved to predict the efficacies of classical drugs, infliximab, vedolizumab, and enterectomy by analyzing huge numbers of clinicopathological variables[154,173-175].
| Ref. | Disease | Aim | Number of samples | ANN technique | Included variables | Outcome |
| Ahmed et al[169], 2017 | CD | Diagnosis | 144 CD patients; 243 HC individuals | BPNN | 103 variables | Accuracy 97.67%; sensitivity 96.07%; specificity 100% |
| Ananthakrishnan et al[154], 2017 | UC and CD | Predicting treatment response to vedolizumab | 43 UC patients; 42 CD patients | vedoNet | Gut microbiome | AUC of CD 88.1%; AUC of UC 85.3% |
| Anekboon et al[201], 2014 | CD | Predicting single nucleotide polymorphisms | 144 CD patients; 243 HC individuals | Multi-layer perceptron network | 103 SNPs | Accuracy 90.4%; sensitivity 87.5%; specificity 92.2% |
| Dong et al[173], 2019 | CD | Predicting the risk of surgical intervention in Chinese patients | 83 patients with surgery; 83 patients without surgery | ANN | 131 variables | Accuracy 90.89%; precision 46.83%; F1 score 0.5757 |
| Fioravanti et al[202], 2018 | IBD | Classification of metagenomics data | 222 IBD patients; 38 HC individuals | CNN | Gut microbiota | - |
| Hardalaç et al[203], 2015 | IBD | Predicting the effect of azathioprine on mucosal healing | 129 IBD patients | BPNN | Age, age at diagnosis, usage of other medications prior to azathioprine use, smoking, sex, UC-CD | Accuracy 79.1% |
| Kirchberger-Tolstik et al[170], 2020 | UC | Diagnosis | 227 Raman maps with 567500 spectra | CNN | Images of Raman spectroscopy | sensitivity of 78%; specificity 93% |
| Klein et al[204], 2017 | CD | Predicting the clinical phenotype | 47 B1 patients; 19 B2 patients; 39 B3 patients | Two-layer FNN | H&E | B1 vs B2 phenotype: sensitivity 81%, specificity 74%, accuracy 75%, AUC 0.74; B1 vs B3 phenotype: sensitivity 69%, specificity 76%, accuracy 70.5%, AUC 0.78; B2 vs B3 phenotype: sensitivity 67%, specificity 72.5%, accuracy 69%, AUC 0.72 |
| Lamash et al[71], 2019 | CD | Visualization and quantitative estimation of CD | 23 pediatric CD patients | CNN | MRI | DSCs of 75 ± 18%, 81 ± 8%, and 97 ± 2% for the lumen, wall, and background, respectively |
| Le et al[174], 2020 | IBD | Predicting IBD and treatment status | 68 CD patients; 53 UC patients; 34 HC individuals | Neural encoder-decoder (NED) network | Gut microbiota | CD vs HC: 95.2% AUC; UC vs HC: 92.5% AUC; CD vs UC: 81.8% AUC |
| Morilla et al[175], 2019 | UC | Predicting treatment responses to infliximab for patients with acute severe UC | 47 patients with acute severe ulcerative colitis | Deep neural network | MicroRNA profiles | 84% accuracy; 0.82 AUC |
| Ozawa et al[112], 2019 | UC | Identification of endoscopic inflammation severity | 841 patients | CNN (GoogLeNet) | Colonoscopy images | 0.86 AUC of Mayo 0; 0.98 AUC of Mayo 0-1 |
| Peng et al[205], 2015 | IBD | Predicting the frequency of relapse | 569 UC patients; 332 CD patients | ANN | Meteorological data | High accuracy in predicting the frequency of relapse of IBD (MSE = 0.009, MAPE = 17.1 %) |
| Shepherd et al[171], 2014 | IBD | Differential diagnosis between IBD and IBS | 59 UC patients; 42 CD patients; 34 IBS patients; 46 HC individuals | Multi-layer perceptron neural network | Gas chromatograph coupled to a metal oxide sensor in stool samples | 76% sensitivity, 88% specificity, 76% accuracy |
| Takayama et al[132], 2015 | UC | Predicting treatment response to cytoapheresis | 90 UC patients | Multi-layer perceptron neural network | 13 clinical variables | 96% sensitivity; 97% sensitivity |
| Tong et al[172], 2020 | CD, UC and ITB | Differential diagnosis between CD, UC and ITB | 5128 UC patients; 875 CD patients; ITB 396 patients | CNN | Differential features of endoscopic images between UC, CD and ITB | The precisions/recalls of UC-CD-ITB when employing the CNN were 0.99/0.97, 0.87/0.83, and 0.52/0.81, respectively |
Moreover, ANNs exhibited remarkable detective abilities for GI bleeding and atrophic gastritis. These two distinct diseases have one similarity: Early detection has critical value while delayed diagnosis can lead to a poor prognosis. However, the rates of missed diagnosis for GI bleeding and atrophic gastritis are relatively high. ANNs may overcome this dilemma. According to the existing evidence, the detection accuracies of endoscopy were significantly elevated under the assistance of ANN models[111,176-178]. Collectively, these studies indicate the role of ANN in the clinical practice of GI noncancerous diseases.
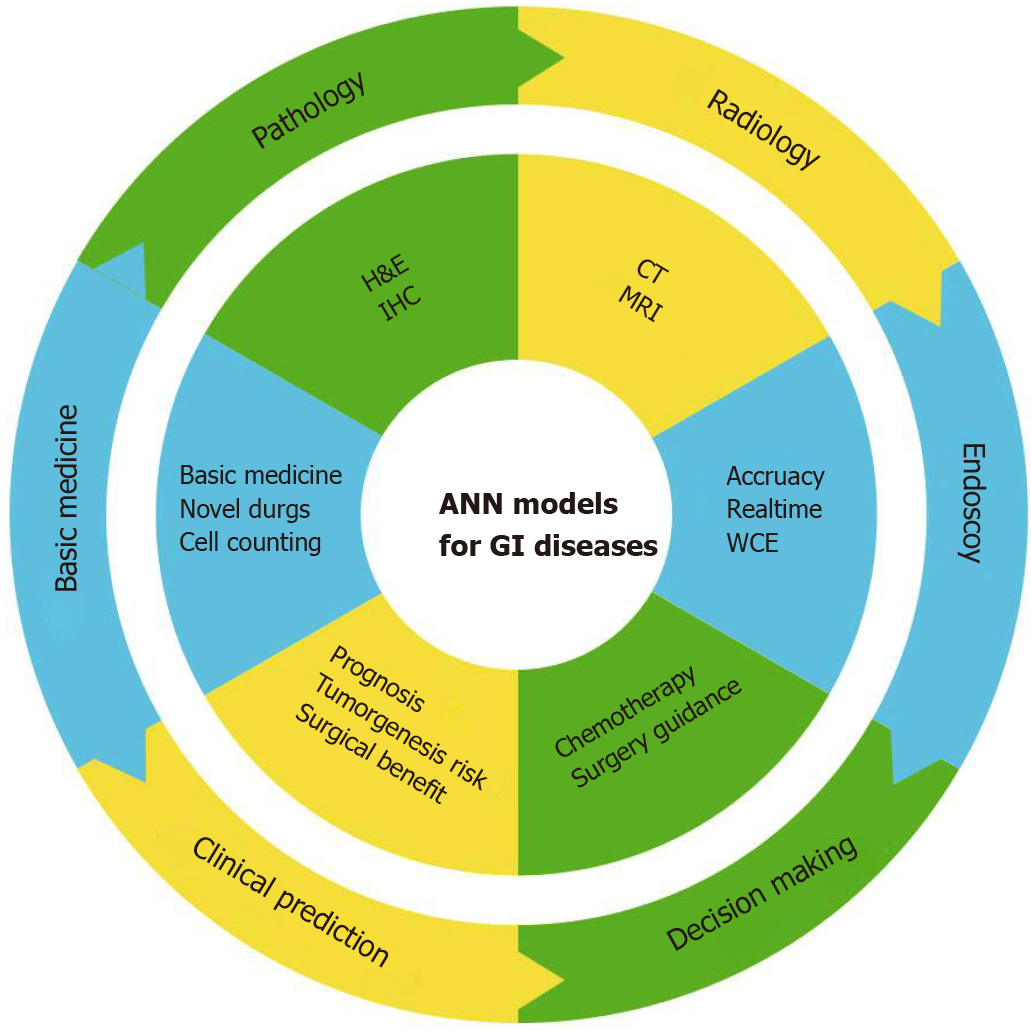
As mentioned above, accumulating studies have demonstrated that ANNs might have remarkable potential in diagnosis and treatment of GI diseases (Figure 3). ANN-based models commonly have optimal accuracies and AUC values. Some evaluation indexes of ANN models even achieved an accuracy of 100%[81,179]. For further validation of ANN metrics, comparisons were also performed and can be divided into three aspects based on the compared objects. First, regarding experts, studies involving comparisons with experts aimed at showing ANN superiority over artificial diagnosis[8,86,136]. The majority of ANNs exhibited faster speed and better accuracy than expert clinicians. Therefore, certain authors claimed that ANNs would become excellent tools for clinicians and scientists, perhaps even replacing humans in this capacity. Second, different ANN models have been studied. Researchers used several ANN algorithms that were trained with the same dataset and underwent validation phases [144,150,180]. The models with the best fitting degree were then selected out to process for further exploration. The comparisons exhibited the capabilities of different models in handling with clinical problems. Last, other reported data have shown that literature learning is another approach to determine the efficiency of developed models[106,181]. However, this type of comparative method is not reliable due to the inconsistency of research baselines, such as differences in datasets, hardware performance, and running time. These undefined bias may also affect comparative results. This method is thus not recommended for follow-up studies to further verify the advantages of their models. Generally, compelling evidence indicated that ANNs can lead the classification and deep analysis of GI clinical practice compared to linear statistical models and human labor.
Computers outperform humans in data processing, as shown in medical studies. The comprehensive diagnosis of GI diseases requires many types of examinations. GI clinicians must integrate clinical indicators to make diagnosis. However, existing guidelines and expert consensus cannot help GI clinicians manage the majority of complex diseases. ANNs can mitigate this limitation. Many types of patient characteristics and clinical indicators can be included in predictive models. Data stored in pictures is geometrically above that of textual data. CNNs can interpret radiological and endoscopic images. The analytic speed of CNNs is even fast enough to make real-time detection achievable. Some studies simultaneously included clinical indicators and picture information to analyze patient situations. This novel combination exceeds the limitations of human capacity and provides optimal diagnostic and predictive accuracies.
Cost is also an important factor for clinical practice. Cultivating a qualified doctor demands considerable time and economic and social costs. However, ANN learning is a relatively simple process. Based on current reports, training ANN models with large-sample databases requires only several days[20]. ANNs can comprehensively analyze data features and adjust the weight values within an extremely short time. Furthermore, hardware is the basic requirement to run ANN algorithms, specifically graphics cards and central processing units, the two important data-processing elements. High-performance hardware costs a few thousand dollars, which is far cheaper than training a doctor. Collectively, the time and economic cost of training ANN models is much smaller than that of training doctors. It is believed that ANN will play a critical role in clinical practice in the near future.
The application of ANNs appears to be a way to overcome existing problems for GI doctors. However, certain limitations should be taken seriously. First, training methods affect the efficiency of ANN models. Most research groups have performed small-sample studies with fewer than 1000 cases. For endoscopic detection, although the number of freezing frames could reach ten thousand, some were from the same video[8,182]. Thus, frame repeatability impaired their representativeness. The small size of dataset led to a higher likelihood of overfitting, which refers to models that receive too much training and overly fit the characteristics of internal datasets. Rigid observation of internal datasets will naturally reduce the model’s flexibility to manage external data. For example, if the model is trained with a dataset of patients from country A, patients from country B may not be well described by this model due to various factors. Otherwise, retrospective datasets should be used as the primary sources of ANN training. Similar to common retrospective studies, biases also exist and affect the application of models. Selection bias is the most important factor. Patient data from medical histories could be selected based on research programmes. However, some studies used public databases to train their models[167,183]. The information of registered cases is inflexible, which means that researchers cannot exclude potential bias as they expected.
Second, the compromise between interpretability and accuracy should be emphasized. Models with optimal accuracy typically lack interpretability (i.e., explainability). Conversely, models with nearly perfect interpretability are less accurate, like linear regression. During ANN training, algorithm modulation and data feedback are blind. Researchers do not know the activation functions in hidden layers, which are similar to a machine that hides its circuit in the box and places only switches and displayers outside; this feature is called as “black box”. For important medical decisions, doctors and patients both have the right to know how decisions were made. An inability to understand how the model makes decisions will naturally reduce the patients’ and doctors’ confidence. Conversely, lack of explainability makes researchers unable to describe the deduction processes of ANN models, decreasing their referential value.
Third, sociological issues should be carefully considered for ANN applications, which could be divided into three aspects. First, regarding medical liability, mistakes and medical negligence are likely, despite the outstanding ANN performance in the validation phase. Thus, who should be responsible for decisions and how to distribute liability to pay for compensation are difficult to determine. Doctors, patients, and programmers are all seemingly innocent, and accidents primarily result from inherent shortcomings, not human carelessness. Second, regarding confidentiality, the operation of ANN models requires many types of data, some of which concern patient privacy. Doctors who betray privacies will receive punishment. However, ANN models are connected to the Internet or company intranets. It is difficult to distinguish the origins of a privacy disclosure. Furthermore, the capabilities of data extraction by ANNs are incomparable. The ranges of consent exemption thus require further argument. The balance between ANN research and patient privacy remains a challenge. Last, regarding legislation, multiparty participation is an important characteristic of ANNs, which should include hospitals, patients, product providers, and insurance companies. Regretfully, the rules of ANNs used in medical practice are still obscure. Existing legal norms cannot support the application and prospective studies of ANNs.
The goal of ANN applications is to maximize medical benefits for patients, doctors, and society. To provide GI diagnosis and recommended treatment, ANNs must be further improved to bring them in line with clinical standards and inherent characteristics. Future directions are described below.
Enlargement of application ranges: ANNs have been used for GI diagnosis, prognosis and risk prediction, and therapeutic guidance, achieving relatively high accuracies. Significance is limited to constructing more models that have been proposed in the same fields[184,185]. Future studies should focus on expanding ANN application ranges. Treatment decisions and surgical guidance are two representative aspects of future development. They require including more factors and presupposing more complex situations. ANNs may become a promising tool to overcome the difficulties. Epidemiology is another field burdened with large quantities of data. Training ANN-based models will help investigators have a deep and quick command of epidemiologic features.
Development of unsupervised learning: Supervised learning is the mainstream direction of medical research. ANN models are primarily based on labelled datasets, which require considerable time and manpower. Future studies should try to use unsupervised and semi-supervised learning to reduce the training cost and develop comprehensive decision-making models. The unique advantages of unsupervised learning can break classical thoughts of clinical trials. It can automatically reveal the internal structures of mass data without grouping and statistical analysis. Also, unsupervised learning is “clear box” and can produce visual information that can be comprehended by humans, providing more convenient parameter adjustment and systematic optimization. Unsupervised ANN-based models should be created for data pre-treatment and association screening. Mature unsupervised systems for basic research are also required to find more diagnostic biomarkers and therapeutic targets.
Improvement of datasets: Large-sample prospective studies and real-world studies are encouraged. However, these studies will require high-quality datasets, including normalized inclusion criteria, number of cases, detailed degree, and rigorous check. Optimal sources of data can reduce the bias of ANN systems. Otherwise, larger sample sizes do not mean that enrolled cases should cover as many races and countries as possible. Heterogeneity is ubiquitous in medical care. There remains a lack of evidence that ANNs, perhaps even all AI algorithms, have the potential to generalize disease features throughout the world. It should be temporarily accepted to construct ANN models with functional, regional, or subpopulation characteristics rather than wide application. The qualities of the datasets determine the performance of the constructed models. More efforts should be paid to build full equipped registry systems and provide complete data for model training, which is typically neglected by constructors.
Emphasis on legislation: The nature of AI is totally different from human intelligence. It cannot accept liability by its own. Decision-making methods are still based on learning and experience. Also, ANNs are unable to correctly address problems that are beyond their knowledge. That is why commercial AI products are slightly clumsy when managing complex situations. The “black box” feature of ANNs means that all participants are precluded from the decision-making process. Once ANN models make mistakes, how to rationally distribute responsibilities should be carefully answered by legislation. The absence of reasonable rules also slowed clinical trials. Judgment criteria must be outlined to allow clinical trials to be launched. Problems that emerge will then guide legislation. Positive feedback will promote the progress of ANN application.
Progress in ethics: ANNs have been created to solve the specific problems, which naturally forms result-oriented operation patterns. However, this process is not always suitable for making decisions independently, particularly for GI diseases. For example, for an old patient who had severe ulcerative colitis and underwent many partial enterectomies, ANN models may show that he can benefit from the next surgery. Conversely, doctors will persuade patients to accept expectant treatment due to their tolerance, age, and emotion. The absence of empathy will make an ANN’s decision “perfect” but impersonal, which is partially why ANNs cannot become a substitute for human decision makings. Regarding the substitutability of human labor, arguments on this topic have never been stopped. ANNs have gradually taken over simple tasks. Similar to the aforementioned statement, many models outperformed experts in endoscopy, radiology, pathology, and gastroenterology[8,19,56,186]. Then, concerns about ANNs taking jobs from gastroenterologists in the future become more common. We hold the opposite view: The development of ANN techniques can reverse the transmission of pressure to improve the competence of gastroenterologists. Only in this way can pressure and injustice fundamentally be solved. Some issues of ethics and society will be answered by further application of ANNs.
Avoidance of overestimation: A misleading saying is that ANNs automatically produce analytic results after being fed raw data. Many studies emphasize construction methods but neglect the importance of data cleaning and conversion. Insufficient data pre-processing has serious impacts on the qualities of models. Rigorous criteria of data pre-processing should thus be developed. Coincidentally, ANNs have advantages regarding data labelling and classification. ANN models should be developed to help construct other ANN models for decision making. Although results showed that ANNs outperformed other AI methods, certain studies also highlighted their limitations[187,188]. An internal comparison between different ML methods is required.
The relationship between AI and medicine has received much attention. In this review, we introduced the basic knowledge of ANN, a primary method of AI and its application in the diagnosis, risk prediction and treatment of GI diseases. Considering interdisciplinary difficulties, the contents of this review were written from the perspective of gastroenterologists. Many vivid metaphors and brief conclusions reduced the comprehension threshold of GI clinicians and scientists. The characteristics, current limitations, and future directions were then illustrated. Undoubtedly, ANNs provide excellent value in clinical practice and even outperform GI experts and other types of AI techniques. However, there are still certain problems that hinder ANN applications, which require additional exploration in many fields. It is believed that ANN will become one of the most efficient tools for GI clinicians and benefit all participants in the future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jiménez Pérez M, Lalmuanawma S S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Sotos JG. MYCIN and NEOMYCIN: two approaches to generating explanations in rule-based expert systems. Aviat Space Environ Med. 1990;61:950-954. [PubMed] [Cited in This Article: ] |
| 2. | Mungle T, Tewary S, Das DK, Arun I, Basak B, Agarwal S, Ahmed R, Chatterjee S, Chakraborty C. MRF-ANN: a machine learning approach for automated ER scoring of breast cancer immunohistochemical images. J Microsc. 2017;267:117-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Wang JL, Jin GL, Yuan ZG. Artificial neural network predicts hemorrhagic contusions following decompressive craniotomy in traumatic brain injury. J Neurosurg Sci. 2021;65:69-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Zhang Y, van der Werf NR, Jiang B, van Hamersvelt R, Greuter MJW, Xie X. Motion-corrected coronary calcium scores by a convolutional neural network: a robotic simulating study. Eur Radiol. 2020;30:1285-1294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Li A, Quan SF, Silva GE, Perfect MM, Roveda JM. A Novel Artificial Neural Network Based Sleep-Disordered Breathing Screening Tool. J Clin Sleep Med. 2018;14:1063-1069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Chen CY, Chen YF, Chen HY, Hung CT, Shi HY. Artificial Neural Network and Cox Regression Models for Predicting Mortality after Hip Fracture Surgery: A Population-Based Comparison. Medicina (Kaunas). 2020;56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 245] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 8. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019; 157: 1044-1054. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 165] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 9. | Ling T, Wu L, Fu Y, Xu Q, An P, Zhang J, Hu S, Chen Y, He X, Wang J, Chen X, Zhou J, Xu Y, Zou X, Yu H. A deep learning-based system for identifying differentiation status and delineating the margins of early gastric cancer in magnifying narrow-band imaging endoscopy. Endoscopy. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 10. | He K, Zhang X, Ren S, Sun J. Identity Mappings in Deep Residual Networks. In: Leibe B, Matas J, Sebe N, Welling M (eds). Computer Vision – ECCV 2016. Lecture Notes in Computer Science, vol 9908. Springer: Cham, 2016: 630-645. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2134] [Cited by in F6Publishing: 2096] [Article Influence: 262.0] [Reference Citation Analysis (0)] |
| 11. | Fu J, Zhong X, Li N, Van Dams R, Lewis J, Sung K, Raldow AC, Jin J, Qi XS. Deep learning-based radiomic features for improving neoadjuvant chemoradiation response prediction in locally advanced rectal cancer. Phys Med Biol. 2020;65:075001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 12. | Karakitsos P, Ioakim-Liossi A, Pouliakis A, Botsoli-Stergiou EM, Tzivras M, Archimandritis A, Kyrkou K. A comparative study of three variations of the learning vector quantizer in the discrimination of benign from malignant gastric cells. Cytopathology. 1998;9:114-125. [PubMed] [Cited in This Article: ] |
| 13. | Song Z, Zou S, Zhou W, Huang Y, Shao L, Yuan J, Gou X, Jin W, Wang Z, Chen X, Ding X, Liu J, Yu C, Ku C, Liu C, Sun Z, Xu G, Wang Y, Zhang X, Wang D, Wang S, Xu W, Davis RC, Shi H. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11:4294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 14. | Kosaraju SC, Hao J, Koh HM, Kang M. Deep-Hipo: Multi-scale receptive field deep learning for histopathological image analysis. Methods. 2020;179:3-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Qu J, Hiruta N, Terai K, Nosato H, Murakawa M, Sakanashi H. Gastric Pathology Image Classification Using Stepwise Fine-Tuning for Deep Neural Networks. J Healthc Eng. 2018;2018:8961781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 16. | Liang Q, Nan Y, Coppola G, Zou K, Sun W, Zhang D, Wang Y, Yu G. Weakly Supervised Biomedical Image Segmentation by Reiterative Learning. IEEE J Biomed Health Inform. 2019;23:1205-1214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 17. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 18. | Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, Higgins J, Rubin DL, Shen J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22:132-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 162] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 19. | Wei JW, Suriawinata AA, Vaickus LJ, Ren B, Liu X, Lisovsky M, Tomita N, Abdollahi B, Kim AS, Snover DC, Baron JA, Barry EL, Hassanpour S. Evaluation of a Deep Neural Network for Automated Classification of Colorectal Polyps on Histopathologic Slides. JAMA Netw Open. 2020;3:e203398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Yoon H, Lee J, Oh JE, Kim HR, Lee S, Chang HJ, Sohn DK. Tumor Identification in Colorectal Histology Images Using a Convolutional Neural Network. J Digit Imaging. 2019;32:131-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Graham S, Chen H, Gamper J, Dou Q, Heng PA, Snead D, Tsang YW, Rajpoot N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med Image Anal. 2019;52:199-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 22. | Graham S, Vu QD, Raza SEA, Azam A, Tsang YW, Kwak JT, Rajpoot N. Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58:101563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 305] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 23. | Zhang S, Han F, Liang Z, Tan J, Cao W, Gao Y, Pomeroy M, Ng K, Hou W. An investigation of CNN models for differentiating malignant from benign lesions using small pathologically proven datasets. Comput Med Imaging Graph. 2019;77:101645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Sena P, Fioresi R, Faglioni F, Losi L, Faglioni G, Roncucci L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol Lett. 2019;18:6101-6107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Wang KS, Yu G, Xu C, Meng XH, Zhou J, Zheng C, Deng Z, Shang L, Liu R, Su S, Zhou X, Li Q, Li J, Wang J, Ma K, Qi J, Hu Z, Tang P, Deng J, Qiu X, Li BY, Shen WD, Quan RP, Yang JT, Huang LY, Xiao Y, Yang ZC, Li Z, Wang SC, Ren H, Liang C, Guo W, Li Y, Xiao H, Gu Y, Yun JP, Huang D, Song Z, Fan X, Chen L, Yan X, Huang ZC, Huang J, Luttrell J, Zhang CY, Zhou W, Zhang K, Yi C, Wu C, Shen H, Wang YP, Xiao HM, Deng HW. Accurate diagnosis of colorectal cancer based on histopathology images using artificial intelligence. BMC Med. 2021;19:76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 26. | Awan R, Sirinukunwattana K, Epstein D, Jefferyes S, Qidwai U, Aftab Z, Mujeeb I, Snead D, Rajpoot N. Glandular Morphometrics for Objective Grading of Colorectal Adenocarcinoma Histology Images. Sci Rep. 2017;7:16852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 27. | Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ. 2017;5:e3874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 28. | Xu Y, Li Y, Wang Y, Liu M, Fan Y, Lai M, Chang EI. Gland Instance Segmentation Using Deep Multichannel Neural Networks. IEEE Trans Biomed Eng. 2017;64:2901-2912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Van Eycke YR, Balsat C, Verset L, Debeir O, Salmon I, Decaestecker C. Segmentation of glandular epithelium in colorectal tumours to automatically compartmentalise IHC biomarker quantification: A deep learning approach. Med Image Anal. 2018;49:35-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 297] [Cited by in F6Publishing: 213] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 31. | Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging. 2016;35:1196-1206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 784] [Cited by in F6Publishing: 457] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 32. | Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley CM, Wu CH, Beggs AD, McDermott U, Dunne PD, Meade A, Walker SM, Murray GI, Samuel L, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH; S:CORT consortium. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544-554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 108] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 33. | Matsumoto T, Niioka H, Kumamoto Y, Sato J, Inamori O, Nakao R, Harada Y, Konishi E, Otsuji E, Tanaka H, Miyake J, Takamatsu T. Deep-UV excitation fluorescence microscopy for detection of lymph node metastasis using deep neural network. Sci Rep. 2019;9:16912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Shiraishi T, Shinto E, Nearchou IP, Tsuda H, Kajiwara Y, Einama T, Caie PD, Kishi Y, Ueno H. Prognostic significance of mesothelin expression in colorectal cancer disclosed by area-specific four-point tissue microarrays. Virchows Arch. 2020;477:409-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Luo XJ, Zhang B, Li JG, Luo XA, Yang LF. Autofluorescence spectroscopy for evaluating dysplasia in colorectal tissues. Z Med Phys. 2012;22:40-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 36. | Jia Z, Huang X, Chang EI, Xu Y. Constrained Deep Weak Supervision for Histopathology Image Segmentation. IEEE Trans Med Imaging. 2017;36:2376-2388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 37. | Qaiser T, Tsang YW, Taniyama D, Sakamoto N, Nakane K, Epstein D, Rajpoot N. Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Med Image Anal. 2019;55:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 38. | Su H, Ruan J, Chen T, Lin E, Shi L. CT-assessed sarcopenia is a predictive factor for both long-term and short-term outcomes in gastrointestinal oncology patients: a systematic review and meta-analysis. Cancer Imaging. 2019;19:82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 39. | Umezawa S, Nagata N, Arimoto J, Uchiyama S, Higurashi T, Nakano K, Ishii N, Sakurai T, Moriyasu S, Takeda Y, Nagase H, Komatsu H, Nakajima A, Mizuki A. Contrast-enhanced CT for Colonic Diverticular Bleeding before Colonoscopy: A Prospective Multicenter Study. Radiology. 2018;288:755-761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Mehdy MM, Ng PY, Shair EF, Saleh NIM, Gomes C. Artificial Neural Networks in Image Processing for Early Detection of Breast Cancer. Comput Math Methods Med. 2017;2017:2610628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 41. | Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Weinreb JC, Duncan JS, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29:3348-3357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 42. | Knoll P, Mirzaei S, Müllner A, Leitha T, Koriska K, Köhn H, Neumann M. An artificial neural net and error backpropagation to reconstruct single photon emission computerized tomography data. Med Phys. 1999;26:244-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 43. | Zheng L, Zhang X, Hu J, Gao Y, Zhang M, Li S, Zhou X, Niu T, Lu Y, Wang D. Establishment and Applicability of a Diagnostic System for Advanced Gastric Cancer T Staging Based on a Faster Region-Based Convolutional Neural Network. Front Oncol. 2020;10:1238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Suzuki K, Rockey DC, Dachman AH. CT colonography: advanced computer-aided detection scheme utilizing MTANNs for detection of "missed" polyps in a multicenter clinical trial. Med Phys. 2010;37:12-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 45. | Suzuki K, Zhang J, Xu J. Massive-training artificial neural network coupled with Laplacian-eigenfunction-based dimensionality reduction for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging. 2010;29:1907-1917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 46. | Liu X, Guo S, Zhang H, He K, Mu S, Guo Y, Li X. Accurate colorectal tumor segmentation for CT scans based on the label assignment generative adversarial network. Med Phys. 2019;46:3532-3542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 47. | Chen Y, Ren Y, Fu L, Xiong J, Larsson R, Xu X, Sun J, Zhao J. A 3D Convolutional Neural Network Framework for Polyp Candidates Detection on the Limited Dataset of CT Colonography. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:678-681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Jerebko AK, Malley JD, Franaszek M, Summers RM. Multiple neural network classification scheme for detection of colonic polyps in CT colonography data sets. Acad Radiol. 2003;10:154-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 49. | Jerebko AK, Summers RM, Malley JD, Franaszek M, Johnson CD. Computer-assisted detection of colonic polyps with CT colonography using neural networks and binary classification trees. Med Phys. 2003;30:52-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 50. | Suzuki K, Yoshida H, Näppi J, Dachman AH. Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes. Med Phys. 2006;33:3814-3824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Men K, Boimel P, Janopaul-Naylor J, Zhong H, Huang M, Geng H, Cheng C, Fan Y, Plastaras JP, Ben-Josef E, Xiao Y. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol. 2018;63:185016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers RM. Improving Computer-Aided Detection Using Convolutional Neural Networks and Random View Aggregation. IEEE Trans Med Imaging. 2016;35:1170-1181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 411] [Cited by in F6Publishing: 256] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 53. | Gao Y, Zhang ZD, Li S, Guo YT, Wu QY, Liu SH, Yang SJ, Ding L, Zhao BC, Lu Y. Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer. Chin Med J (Engl). 2019;132:2804-2811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 54. | Huang Z, Liu D, Chen X, Yu P, Wu J, Song B, Hu J, Wu B. Retrospective imaging studies of gastric cancer: Study protocol clinical trial (SPIRIT Compliant). Medicine (Baltimore). 2020;99:e19157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 55. | Hou Z, Yang Y, Li S, Yan J, Ren W, Liu J, Wang K, Liu B, Wan S. Radiomic analysis using contrast-enhanced CT: predict treatment response to pulsed low dose rate radiotherapy in gastric carcinoma with abdominal cavity metastasis. Quant Imaging Med Surg. 2018;8:410-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Khalili K, Lawlor RL, Pourafkari M, Lu H, Tyrrell P, Kim TK, Jang HJ, Johnson SA, Martel AL. Convolutional neural networks versus radiologists in characterization of small hypoattenuating hepatic nodules on CT: a critical diagnostic challenge in staging of colorectal carcinoma. Sci Rep. 2020;10:15248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 57. | Tan J, Gao Y, Liang Z, Cao W, Pomeroy MJ, Huo Y, Li L, Barish MA, Abbasi AF, Pickhardt PJ. 3D-GLCM CNN: A 3-Dimensional Gray-Level Co-Occurrence Matrix-Based CNN Model for Polyp Classification via CT Colonography. IEEE Trans Med Imaging. 2020;39:2013-2024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 58. | Trebeschi S, van Griethuysen JJM, Lambregts DMJ, Lahaye MJ, Parmar C, Bakers FCH, Peters NHGM, Beets-Tan RGH, Aerts HJWL. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci Rep. 2017;7:5301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 59. | Panic J, Defeudis A, Mazzetti S, Rosati S, Giannetto G, Vassallo L, Regge D, Balestra G, Giannini V. A Convolutional Neural Network based system for Colorectal cancer segmentation on MRI images. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:1675-1678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 60. | Wang J, Lu J, Qin G, Shen L, Sun Y, Ying H, Zhang Z, Hu W. Technical Note: A deep learning-based autosegmentation of rectal tumors in MR images. Med Phys. 2018;45:2560-2564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 61. | Wang M, Xie P, Ran Z, Jian J, Zhang R, Xia W, Yu T, Ni C, Gu J, Gao X, Meng X. Full convolutional network based multiple side-output fusion architecture for the segmentation of rectal tumors in magnetic resonance images: A multi-vendor study. Med Phys. 2019;46:2659-2668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 62. | Soomro MH, Coppotelli M, Conforto S, Schmid M, Giunta G, Del Secco L, Neri E, Caruso D, Rengo M, Laghi A. Automated Segmentation of Colorectal Tumor in 3D MRI Using 3D Multiscale Densely Connected Convolutional Neural Network. J Healthc Eng. 2019;2019:1075434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 63. | Wang D, Xu J, Zhang Z, Li S, Zhang X, Zhou Y, Lu Y. Evaluation of Rectal Cancer Circumferential Resection Margin Using Faster Region-Based Convolutional Neural Network in High-Resolution Magnetic Resonance Images. Dis Colon Rectum. 2020;63:143-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 64. | Shi L, Zhang Y, Nie K, Sun X, Niu T, Yue N, Kwong T, Chang P, Chow D, Chen JH, Su MY. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn Reson Imaging. 2019;61:33-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 65. | Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, Niu T, Sun X. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin Cancer Res. 2016;22:5256-5264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 281] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 66. | Bangi E, Ang C, Smibert P, Uzilov AV, Teague AG, Antipin Y, Chen R, Hecht C, Gruszczynski N, Yon WJ, Malyshev D, Laspina D, Selkridge I, Rainey H, Moe AS, Lau CY, Taik P, Wilck E, Bhardwaj A, Sung M, Kim S, Yum K, Sebra R, Donovan M, Misiukiewicz K, Schadt EE, Posner MR, Cagan RL. A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci Adv. 2019;5:eaav6528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 67. | Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, Zagonel V, Leone F, Depetris I, Martinelli E, Troiani T, Ciardiello F, Racca P, Bertotti A, Siravegna G, Torri V, Amatu A, Ghezzi S, Marrapese G, Palmeri L, Valtorta E, Cassingena A, Lauricella C, Vanzulli A, Regge D, Veronese S, Comoglio PM, Bardelli A, Marsoni S, Siena S. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738-746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 549] [Cited by in F6Publishing: 644] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 68. | He K, Liu X, Li M, Li X, Yang H, Zhang H. Noninvasive KRAS mutation estimation in colorectal cancer using a deep learning method based on CT imaging. BMC Med Imaging. 2020;20:59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 69. | Wu X, Li Y, Chen X, Huang Y, He L, Zhao K, Huang X, Zhang W, Dong M, Huang J, Xia T, Liang C, Liu Z. Deep Learning Features Improve the Performance of a Radiomics Signature for Predicting KRAS Status in Patients with Colorectal Cancer. Acad Radiol. 2020;27:e254-e262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 70. | Huang L, Li M, Gou S, Zhang X, Jiang K. Automated Segmentation Method for Low Field 3D Stomach MRI Using Transferred Learning Image Enhancement Network. Biomed Res Int. 2021;2021:6679603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Lamash Y, Kurugol S, Freiman M, Perez-Rossello JM, Callahan MJ, Bousvaros A, Warfield SK. Curved planar reformatting and convolutional neural network-based segmentation of the small bowel for visualization and quantitative assessment of pediatric Crohn's disease from MRI. J Magn Reson Imaging. 2019;49:1565-1576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 72. | Laursen SB, Leontiadis GI, Stanley AJ, Møller MH, Hansen JM, Schaffalitzky de Muckadell OB. Relationship between timing of endoscopy and mortality in patients with peptic ulcer bleeding: a nationwide cohort study. Gastrointest Endosc 2017; 85: 936-944. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 81] [Article Influence: 11.6] [Reference Citation Analysis (1)] |
| 73. | Ogura M, Hikiba Y, Maeda S, Matsumura M, Okano K, Sassa R, Yoshida H, Kawabe T, Omata M. Mortality from gastric cancer in patients followed with upper gastrointestinal endoscopy. Scand J Gastroenterol. 2008;43:574-580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 74. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 373] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 75. | Yoon HJ, Kim S, Kim JH, Keum JS, Oh SI, Jo J, Chun J, Youn YH, Park H, Kwon IG, Choi SH, Noh SH. A Lesion-Based Convolutional Neural Network Improves Endoscopic Detection and Depth Prediction of Early Gastric Cancer. J Clin Med. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 76. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc 2019; 89: 806-815. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 195] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 77. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 78. | Gao J, Guo Y, Sun Y, Qu G. Application of Deep Learning for Early Screening of Colorectal Precancerous Lesions under White Light Endoscopy. Comput Math Methods Med. 2020;2020:8374317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 79. | Bagheri M, Mohrekesh M, Tehrani M, Najarian K, Karimi N, Samavi S, Reza Soroushmehr SM. Deep Neural Network based Polyp Segmentation in Colonoscopy Images using a Combination of Color Spaces. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:6742-6745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 80. | Zhang X, Hu W, Chen F, Liu J, Yang Y, Wang L, Duan H, Si J. Gastric precancerous diseases classification using CNN with a concise model. PLoS One. 2017;12:e0185508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 81. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 82. | Zhang R, Zheng Y, Poon CCY, Shen D, Lau JYW. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recognit. 2018;83:209-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 83. | Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2020; 92: 144-151. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 91] [Article Influence: 22.8] [Reference Citation Analysis (1)] |
| 84. | Zhang X, Chen F, Yu T, An J, Huang Z, Liu J, Hu W, Wang L, Duan H, Si J. Real-time gastric polyp detection using convolutional neural networks. PLoS One. 2019;14:e0214133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 85. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 209] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 86. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 87. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 82] [Article Influence: 20.5] [Reference Citation Analysis (1)] |
| 88. | Sakai Y, Takemoto S, Hori K, Nishimura M, Ikematsu H, Yano T, Yokota H. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:4138-4141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 89. | Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, Ding Q, Mu G, Yin A, Huang X, Liu J, Jiang X, Wang Z, Deng Y, Liu M, Lin R, Ling T, Li P, Wu Q, Jin P, Chen J, Yu H. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 90. | Hwang M, Wang D, Kong XX, Wang Z, Li J, Jiang WC, Hwang KS, Ding K. An automated detection system for colonoscopy images using a dual encoder-decoder model. Comput Med Imaging Graph. 2020;84:101763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 91. | Wang W, Tian J, Zhang C, Luo Y, Wang X, Li J. An improved deep learning approach and its applications on colonic polyp images detection. BMC Med Imaging. 2020;20:83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 92. | Xu M, Zhou W, Wu L, Zhang J, Wang J, Mu G, Huang X, Li Y, Yuan J, Zeng Z, Wang Y, Huang L, Liu J, Yu H. Artificial intelligence in diagnosis of gastric precancerous conditions by image-enhanced endoscopy: a multicenter, diagnostic study (with video). Gastrointest Endosc. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 93. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 94. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Endoscopic three-categorical diagnosis of Helicobacter pylori infection using linked color imaging and deep learning: a single-center prospective study (with video). Gastric Cancer. 2020;23:1033-1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 95. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 96. | Zorron Cheng Tao Pu L, Maicas G, Tian Y, Yamamura T, Nakamura M, Suzuki H, Singh G, Rana K, Hirooka Y, Burt AD, Fujishiro M, Carneiro G, Singh R. Computer-aided diagnosis for characterization of colorectal lesions: comprehensive software that includes differentiation of serrated lesions. Gastrointest Endosc. 2020;92:891-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 97. | Zheng Y, Zhang R, Yu R, Jiang Y, Mak TWC, Wong SH, Lau JYW, Poon CCY. Localisation of Colorectal Polyps by Convolutional Neural Network Features Learnt from White Light and Narrow Band Endoscopic Images of Multiple Databases. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:4142-4145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 98. | Lai LL, Blakely A, Invernizzi M, Lin J, Kidambi T, Melstrom KA, Yu K, Lu T. Separation of color channels from conventional colonoscopy images improves deep neural network detection of polyps. J Biomed Opt. 2021;26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 99. | Voderholzer WA, Beinhoelzl J, Rogalla P, Murrer S, Schachschal G, Lochs H, Ortner MA. Small bowel involvement in Crohn's disease: a prospective comparison of wireless capsule endoscopy and computed tomography enteroclysis. Gut. 2005;54:369-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 233] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 100. | Fritscher-Ravens A, Scherbakov P, Bufler P, Torroni F, Ruuska T, Nuutinen H, Thomson M, Tabbers M, Milla P. The feasibility of wireless capsule endoscopy in detecting small intestinal pathology in children under the age of 8 years: a multicentre European study. Gut. 2009;58:1467-1472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 101. | Barbosa DJ, Ramos J, Lima CS. Detection of small bowel tumors in capsule endoscopy frames using texture analysis based on the discrete wavelet transform. Annu Int Conf IEEE Eng Med Biol Soc. 2008;2008:3012-3015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 102. | Barbosa DJ, Ramos J, Correia JH, Lima CS. Automatic detection of small bowel tumors in capsule endoscopy based on color curvelet covariance statistical texture descriptors. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:6683-6686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 103. | Gayathri Devi K, Radhakrishnan R. Automatic segmentation of colon in 3D CT images and removal of opacified fluid using cascade feed forward neural network. Comput Math Methods Med. 2015;2015:670739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 104. | Majid A, Khan MA, Yasmin M, Rehman A, Yousafzai A, Tariq U. Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microsc Res Tech. 2020;83:562-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 105. | Hwang Y, Lee HH, Park C, Tama BA, Kim JS, Cheung DY, Chung WC, Cho YS, Lee KM, Choi MG, Lee S, Lee BI. Improved classification and localization approach to small bowel capsule endoscopy using convolutional neural network. Dig Endosc. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 106. | Tan Q, Ma AJ, Deng H, Wong VW, Tse YK, Yip TC, Wong GL, Ching JY, Chan FK, Yuen PC. A Hybrid Residual Network and Long Short-Term Memory Method for Peptic Ulcer Bleeding Mortality Prediction. AMIA Annu Symp Proc. 2018;2018:998-1007. [PubMed] [Cited in This Article: ] |
| 107. | Hajabdollahi M, Esfandiarpoor R, Najarian K, Karimi N, Samavi S, Reza Soroushmehr SM. Low Complexity CNN Structure for Automatic Bleeding Zone Detection in Wireless Capsule Endoscopy Imaging. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:7227-7230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 108. | Hung CY, Lin CH, Chang CS, Li JL, Lee CC. Predicting Gastrointestinal Bleeding Events from Multimodal In-Hospital Electronic Health Records Using Deep Fusion Networks. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:2447-2450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 109. | Li XL, Zhang H, Zhang XL, Liu H, Xie GT. Exploring transfer learning for gastrointestinal bleeding detection on small-size imbalanced endoscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:1994-1997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 110. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 111. | Zhang Y, Li F, Yuan F, Zhang K, Huo L, Dong Z, Lang Y, Zhang Y, Wang M, Gao Z, Qin Z, Shen L. Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Dig Liver Dis. 2020;52:566-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 112. | Ozawa T, Ishihara S, Fujishiro M, Saito H, Kumagai Y, Shichijo S, Aoyama K, Tada T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc 2019; 89: 416-421. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 113. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2019; 89: 357-363. e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 114. | Chien CW, Lee YC, Ma T, Lee TS, Lin YC, Wang W, Lee WJ. The application of artificial neural networks and decision tree model in predicting post-operative complication for gastric cancer patients. Hepatogastroenterology. 2008;55:1140-1145. [PubMed] [Cited in This Article: ] |
| 115. | Wang YH, Nguyen PA, Islam MM, Li YC, Yang HC. Development of Deep Learning Algorithm for Detection of Colorectal Cancer in EHR Data. Stud Health Technol Inform. 2019;264:438-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 10] [Reference Citation Analysis (0)] |
| 116. | Hsieh MH, Sun LM, Lin CL, Hsieh MJ, Sun K, Hsu CY, Chou AK, Kao CH. Development of a Prediction Model for Colorectal Cancer among Patients with Type 2 Diabetes Mellitus Using a Deep Neural Network. J Clin Med. 2018;7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 117. | Chau R, Jenkins MA, Buchanan DD, Ait Ouakrim D, Giles GG, Casey G, Gallinger S, Haile RW, Le Marchand L, Newcomb PA, Lindor NM, Hopper JL, Win AK. Determining the familial risk distribution of colorectal cancer: a data mining approach. Fam Cancer. 2016;15:241-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 118. | Nartowt BJ, Hart GR, Roffman DA, Llor X, Ali I, Muhammad W, Liang Y, Deng J. Scoring colorectal cancer risk with an artificial neural network based on self-reportable personal health data. PLoS One. 2019;14:e0221421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 119. | Nakahira H, Ishihara R, Aoyama K, Kono M, Fukuda H, Shimamoto Y, Nakagawa K, Ohmori M, Iwatsubo T, Iwagami H, Matsuno K, Inoue S, Matsuura N, Shichijo S, Maekawa A, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Matsunaga T, Tada T. Stratification of gastric cancer risk using a deep neural network. JGH Open. 2020;4:466-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 120. | Dadkhah E, Sikaroodi M, Korman L, Hardi R, Baybick J, Hanzel D, Kuehn G, Kuehn T, Gillevet PM. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019;6:e000297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 121. | He B, Zhang Y, Zhou Z, Wang B, Liang Y, Lang J, Lin H, Bing P, Yu L, Sun D, Luo H, Yang J, Tian G. A Neural Network Framework for Predicting the Tissue-of-Origin of 15 Common Cancer Types Based on RNA-Seq Data. Front Bioeng Biotechnol. 2020;8:737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 122. | Xiao Y, Wu J, Lin Z, Zhao X. A semi-supervised deep learning method based on stacked sparse auto-encoder for cancer prediction using RNA-seq data. Comput Methods Programs Biomed. 2018;166:99-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 123. | Hensler K, Waschulzik T, Mönig SP, Maruyama K, Hölscher AH, Bollschweiler E. Quality-assured Efficient Engineering of Feedforward Neural Networks (QUEEN) -- pretherapeutic estimation of lymph node status in patients with gastric carcinoma. Methods Inf Med. 2005;44:647-654. [PubMed] [Cited in This Article: ] |
| 124. | Mekicar J, Omejc M. Preoperative prediction of lymph node status in gastric cancer patients with the help of computer analysis. Dig Surg. 2009;26:256-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 125. | Spelt L, Nilsson J, Andersson R, Andersson B. Artificial neural networks--a method for prediction of survival following liver resection for colorectal cancer metastases. Eur J Surg Oncol. 2013;39:648-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 126. | Jagric T, Potrc S, Jagric T. Prediction of liver metastases after gastric cancer resection with the use of learning vector quantization neural networks. Dig Dis Sci. 2010;55:3252-3261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 127. | Kwak MS, Lee HH, Yang JM, Cha JM, Jeon JW, Yoon JY, Kim HI. Deep Convolutional Neural Network-Based Lymph Node Metastasis Prediction for Colon Cancer Using Histopathological Images. Front Oncol. 2020;10:619803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 128. | Kurokawa S, Arimura Y, Yamamoto H, Adachi Y, Endo T, Sato T, Suga T, Hosokawa M, Shinomura Y, Imai K. Tumour matrilysin expression predicts metastatic potential of stage I (pT1) colon and rectal cancers. Gut. 2005;54:1751-1758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 129. | Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in F6Publishing: 275] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 130. | Gohari MR, Biglarian A, Bakhshi E, Pourhoseingholi MA. Use of an artificial neural network to determine prognostic factors in colorectal cancer patients. Asian Pac J Cancer Prev. 2011;12:1469-1472. [PubMed] [Cited in This Article: ] |
| 131. | Lee J, An JY, Choi MG, Park SH, Kim ST, Lee JH, Sohn TS, Bae JM, Kim S, Lee H, Min BH, Kim JJ, Jeong WK, Choi DI, Kim KM, Kang WK, Kim M, Seo SW. Deep Learning-Based Survival Analysis Identified Associations Between Molecular Subtype and Optimal Adjuvant Treatment of Patients With Gastric Cancer. JCO Clin Cancer Inform. 2018;2:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 132. | Takayama T, Okamoto S, Hisamatsu T, Naganuma M, Matsuoka K, Mizuno S, Bessho R, Hibi T, Kanai T. Computer-Aided Prediction of Long-Term Prognosis of Patients with Ulcerative Colitis after Cytoapheresis Therapy. PLoS One. 2015;10:e0131197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 133. | Li Z, Wu X, Gao X, Shan F, Ying X, Zhang Y, Ji J. Development and validation of an artificial neural network prognostic model after gastrectomy for gastric carcinoma: An international multicenter cohort study. Cancer Med. 2020;9:6205-6215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 134. | Liu MM, Wen L, Liu YJ, Cai Q, Li LT, Cai YM. Application of data mining methods to improve screening for the risk of early gastric cancer. BMC Med Inform Decis Mak. 2018;18:121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 135. | Meier A, Nekolla K, Hewitt LC, Earle S, Yoshikawa T, Oshima T, Miyagi Y, Huss R, Schmidt G, Grabsch HI. Hypothesis-free deep survival learning applied to the tumour microenvironment in gastric cancer. J Pathol Clin Res. 2020;6:273-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 136. | Bottaci L, Drew PJ, Hartley JE, Hadfield MB, Farouk R, Lee PW, Macintyre IM, Duthie GS, Monson JR. Artificial neural networks applied to outcome prediction for colorectal cancer patients in separate institutions. Lancet. 1997;350:469-472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 147] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 137. | Korhani Kangi A, Bahrampour A. Predicting the Survival of Gastric Cancer Patients Using Artificial and Bayesian Neural Networks. Asian Pac J Cancer Prev. 2018;19:487-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 14] [Reference Citation Analysis (0)] |
| 138. | Amiri Z, Mohammad K, Mahmoudi M, Parsaeian M, Zeraati H. Assessing the effect of quantitative and qualitative predictors on gastric cancer individuals survival using hierarchical artificial neural network models. Iran Red Crescent Med J. 2013;15:42-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 139. | Bollschweiler EH, Mönig SP, Hensler K, Baldus SE, Maruyama K, Hölscher AH. Artificial neural network for prediction of lymph node metastases in gastric cancer: a phase II diagnostic study. Ann Surg Oncol. 2004;11:506-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 140. | Altomare DF, Di Lena M, Porcelli F, Trizio L, Travaglio E, Tutino M, Dragonieri S, Memeo V, de Gennaro G. Exhaled volatile organic compounds identify patients with colorectal cancer. Br J Surg. 2013;100:144-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 168] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 141. | Altomare DF, Di Lena M, Porcelli F, Travaglio E, Longobardi F, Tutino M, Depalma N, Tedesco G, Sardaro A, Memeo R, de Gennaro G. Effects of Curative Colorectal Cancer Surgery on Exhaled Volatile Organic Compounds and Potential Implications in Clinical Follow-up. Ann Surg. 2015;262:862-6; discussion 866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 142. | Gemoll T, Strohkamp S, Schillo K, Thorns C, Habermann JK. MALDI-imaging reveals thymosin beta-4 as an independent prognostic marker for colorectal cancer. Oncotarget. 2015;6:43869-43880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 143. | Zhao K, Li Z, Yao S, Wang Y, Wu X, Xu Z, Wu L, Huang Y, Liang C, Liu Z. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. EBioMedicine. 2020;61:103054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 144. | Hang X, Li D, Wang J, Wang G. Prognostic significance of microsatellite instabilityassociated pathways and genes in gastric cancer. Int J Mol Med. 2018;42:149-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 145. | Francis NK, Luther A, Salib E, Allanby L, Messenger D, Allison AS, Smart NJ, Ockrim JB. The use of artificial neural networks to predict delayed discharge and readmission in enhanced recovery following laparoscopic colorectal cancer surgery. Tech Coloproctol. 2015;19:419-428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 146. | Lee SM, Kang JO, Suh YM. Comparison of hospital charge prediction models for colorectal cancer patients: neural network vs. decision tree models. J Korean Med Sci. 2004;19:677-681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 147. | Wang J, Li M, Hu YT, Zhu Y. Comparison of hospital charge prediction models for gastric cancer patients: neural network vs. decision tree models. BMC Health Serv Res. 2009;9:161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 148. | Maslekar S, Gardiner AB, Monson JR, Duthie GS. Artificial neural networks to predict presence of significant pathology in patients presenting to routine colorectal clinics. Colorectal Dis. 2010;12:1254-1259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 149. | Bibault JE, Giraud P, Housset M, Durdux C, Taieb J, Berger A, Coriat R, Chaussade S, Dousset B, Nordlinger B, Burgun A. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 150. | Huang CM, Huang MY, Huang CW, Tsai HL, Su WC, Chang WC, Wang JY, Shi HY. Machine learning for predicting pathological complete response in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy. Sci Rep. 2020;10:12555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 151. | Maaref A, Romero FP, Montagnon E, Cerny M, Nguyen B, Vandenbroucke F, Soucy G, Turcotte S, Tang A, Kadoury S. Predicting the Response to FOLFOX-Based Chemotherapy Regimen from Untreated Liver Metastases on Baseline CT: a Deep Neural Network Approach. J Digit Imaging. 2020;33:937-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 152. | Ahn SH, Yeo AU, Kim KH, Kim C, Goh Y, Cho S, Lee SB, Lim YK, Kim H, Shin D, Kim T, Kim TH, Youn SH, Oh ES, Jeong JH. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat Oncol. 2019;14:213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 153. | Lin YC, Huang WT, Ou SC, Hung HH, Cheng WZ, Lin SS, Lin HJ, Huang ST. Neural network analysis of Chinese herbal medicine prescriptions for patients with colorectal cancer. Complement Ther Med. 2019;42:279-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 154. | Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 2017; 21: 603-610. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 260] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 155. | Hildebrand LA, Pierce CJ, Dennis M, Paracha M, Maoz A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers (Basel). 2021;13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 156. | Kitaguchi D, Takeshita N, Matsuzaki H, Oda T, Watanabe M, Mori K, Kobayashi E, Ito M. Automated laparoscopic colorectal surgery workflow recognition using artificial intelligence: Experimental research. Int J Surg. 2020;79:88-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 157. | Kitaguchi D, Takeshita N, Matsuzaki H, Takano H, Owada Y, Enomoto T, Oda T, Miura H, Yamanashi T, Watanabe M, Sato D, Sugomori Y, Hara S, Ito M. Real-time automatic surgical phase recognition in laparoscopic sigmoidectomy using the convolutional neural network-based deep learning approach. Surg Endosc. 2020;34:4924-4931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 158. | Bidaut G, Stoeckert CJ Jr. Characterization of unknown adult stem cell samples by large scale data integration and artificial neural networks. Pac Symp Biocomput. 2009;356-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 159. | Bloom GC, Eschrich S, Zhou JX, Coppola D, Yeatman TJ. Elucidation of a protein signature discriminating six common types of adenocarcinoma. Int J Cancer. 2007;120:769-775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 160. | Liu Q, Junker A, Murakami K, Hu P. Automated Counting of Cancer Cells by Ensembling Deep Features. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 161. | Xuan P, Pan S, Zhang T, Liu Y, Sun H. Graph Convolutional Network and Convolutional Neural Network Based Method for Predicting lncRNA-Disease Associations. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 162. | Xuan P, Sheng N, Zhang T, Liu Y, Guo Y. CNNDLP: A Method Based on Convolutional Autoencoder and Convolutional Neural Network with Adjacent Edge Attention for Predicting lncRNA-Disease Associations. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 163. | Joo M, Park A, Kim K, Son WJ, Lee HS, Lim G, Lee J, Lee DH, An J, Kim JH, Ahn T, Nam S. A Deep Learning Model for Cell Growth Inhibition IC50 Prediction and Its Application for Gastric Cancer Patients. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 164. | Berishvili VP, Perkin VO, Voronkov AE, Radchenko EV, Syed R, Venkata Ramana Reddy C, Pillay V, Kumar P, Choonara YE, Kamal A, Palyulin VA. Time-Domain Analysis of Molecular Dynamics Trajectories Using Deep Neural Networks: Application to Activity Ranking of Tankyrase Inhibitors. J Chem Inf Model. 2019;59:3519-3532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 165. | Liu W, Gao X, Cai Q, Li J, Zhu Z, Li C, Yao X, Yang Q, Xiang M, Yan M. Identification of novel serum biomarkers for gastric cancer by magnetic bead. Front Biosci (Elite Ed). 2010;2:961-971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 166. | Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ, Kerin MJ. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415-1422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 167. | Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu X, Fu Q, Tong Z, Liu L, Zheng Y, Zhao P, Fang W, Liu F. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunother Cancer. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 168. | Jing J, Ge M, Yang Z, Li P. Spatial distribution characteristics of tumor marker CA724 reference values in China. Cancer Med. 2019;8:4465-4474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 169. | Ahmed SS, Dey N, Ashour AS, Sifaki-Pistolla D, Bălas-Timar D, Balas VE, Tavares JM. Effect of fuzzy partitioning in Crohn's disease classification: a neuro-fuzzy-based approach. Med Biol Eng Comput. 2017;55:101-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 170. | Kirchberger-Tolstik T, Pradhan P, Vieth M, Grunert P, Popp J, Bocklitz TW, Stallmach A. Towards an Interpretable Classifier for Characterization of Endoscopic Mayo Scores in Ulcerative Colitis Using Raman Spectroscopy. Anal Chem. 2020;92:13776-13784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 171. | Shepherd SF, McGuire ND, de Lacy Costello BP, Ewen RJ, Jayasena DH, Vaughan K, Ahmed I, Probert CS, Ratcliffe NM. The use of a gas chromatograph coupled to a metal oxide sensor for rapid assessment of stool samples from irritable bowel syndrome and inflammatory bowel disease patients. J Breath Res. 2014;8:026001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 172. | Tong Y, Lu K, Yang Y, Li J, Lin Y, Wu D, Yang A, Li Y, Yu S, Qian J. Can natural language processing help differentiate inflammatory intestinal diseases in China? BMC Med Inform Decis Mak. 2020;20:248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 173. | Dong Y, Xu L, Fan Y, Xiang P, Gao X, Chen Y, Zhang W, Ge Q. A novel surgical predictive model for Chinese Crohn's disease patients. Medicine (Baltimore). 2019;98:e17510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 174. | Le V, Quinn TP, Tran T, Venkatesh S. Deep in the Bowel: Highly Interpretable Neural Encoder-Decoder Networks Predict Gut Metabolites from Gut Microbiome. BMC Genomics. 2020;21:256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 175. | Morilla I, Uzzan M, Laharie D, Cazals-Hatem D, Denost Q, Daniel F, Belleannee G, Bouhnik Y, Wainrib G, Panis Y, Ogier-Denis E, Treton X. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17:905-913. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 176. | Li B, Meng MQ. Computer-aided detection of bleeding regions for capsule endoscopy images. IEEE Trans Biomed Eng. 2009;56:1032-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 177. | Li B, Meng MQ. Computer-based detection of bleeding and ulcer in wireless capsule endoscopy images by chromaticity moments. Comput Biol Med. 2009;39:141-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 178. | Kanai M, Togo R, Ogawa T, Haseyama M. Chronic atrophic gastritis detection with a convolutional neural network considering stomach regions. World J Gastroenterol. 2020;26:3650-3659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
| 179. | Wuestemann J, Hupfeld S, Kupitz D, Genseke P, Schenke S, Pech M, Kreissl MC, Grosser OS. Analysis of Bone Scans in Various Tumor Entities Using a Deep-Learning-Based Artificial Neural Network Algorithm-Evaluation of Diagnostic Performance. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 180. | Zhu L, Luo W, Su M, Wei H, Wei J, Zhang X, Zou C. Comparison between artificial neural network and Cox regression model in predicting the survival rate of gastric cancer patients. Biomed Rep. 2013;1:757-760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 181. | Billah M, Waheed S. Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method. Biomed Eng Lett. 2018;8:69-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 182. | Ozawa T, Ishihara S, Fujishiro M, Kumagai Y, Shichijo S, Tada T. Automated endoscopic detection and classification of colorectal polyps using convolutional neural networks. Therap Adv Gastroenterol. 2020;13:1756284820910659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 183. | Al-Bahrani R, Agrawal A, Choudhary A. Survivability prediction of colon cancer patients using neural networks. Health Informatics J. 2019;25:878-891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 184. | Mohan BP, Khan SR, Kassab LL, Ponnada S, Chandan S, Ali T, Dulai PS, Adler DG, Kochhar GS. High pooled performance of convolutional neural networks in computer-aided diagnosis of GI ulcers and/or hemorrhage on wireless capsule endoscopy images: a systematic review and meta-analysis. Gastrointest Endosc 2021; 93: 356-364. e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 185. | Aziz M, Fatima R, Dong C, Lee-Smith W, Nawras A. The impact of deep convolutional neural network-based artificial intelligence on colonoscopy outcomes: A systematic review with meta-analysis. J Gastroenterol Hepatol. 2020;35:1676-1683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 186. | Dolgobrodov SG, Moore P, Marshall R, Bittern R, Steele RJ, Cuschieri A. Artificial neural network: predicted vs observed survival in patients with colonic cancer. Dis Colon Rectum. 2007;50:184-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 187. | Cueto-López N, García-Ordás MT, Dávila-Batista V, Moreno V, Aragonés N, Alaiz-Rodríguez R. A comparative study on feature selection for a risk prediction model for colorectal cancer. Comput Methods Programs Biomed. 2019;177:219-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 188. | Zhang B, Liang XL, Gao HY, Ye LS, Wang YG. Models of logistic regression analysis, support vector machine, and back-propagation neural network based on serum tumor markers in colorectal cancer diagnosis. Genet Mol Res. 2016;15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 189. | Coppedè F, Grossi E, Lopomo A, Spisni R, Buscema M, Migliore L. Application of artificial neural networks to link genetic and environmental factors to DNA methylation in colorectal cancer. Epigenomics. 2015;7:175-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 190. | Liu B, Cui Q, Jiang T, Ma S. A combinational feature selection and ensemble neural network method for classification of gene expression data. BMC Bioinformatics. 2004;5:136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 83] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 191. | Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res. 2004;10:8380-8385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 117] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 192. | Hu HP, Niu ZJ, Bai YP, Tan XH. Cancer classification based on gene expression using neural networks. Genet Mol Res. 2015;14:17605-17611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 193. | Ronen J, Hayat S, Akalin A. Evaluation of colorectal cancer subtypes and cell lines using deep learning. Life Sci Alliance. 2019;2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 194. | Bilsland AE, Pugliese A, Liu Y, Revie J, Burns S, McCormick C, Cairney CJ, Bower J, Drysdale M, Narita M, Sadaie M, Keith WN. Identification of a Selective G1-Phase Benzimidazolone Inhibitor by a Senescence-Targeted Virtual Screen Using Artificial Neural Networks. Neoplasia. 2015;17:704-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 195. | Maniruzzaman M, Jahanur Rahman M, Ahammed B, Abedin MM, Suri HS, Biswas M, El-Baz A, Bangeas P, Tsoulfas G, Suri JS. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput Methods Programs Biomed. 2019;176:173-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 196. | Inglese P, McKenzie JS, Mroz A, Kinross J, Veselkov K, Holmes E, Takats Z, Nicholson JK, Glen RC. Deep learning and 3D-DESI imaging reveal the hidden metabolic heterogeneity of cancer. Chem Sci. 2017;8:3500-3511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 197. | Shi R, Chen W, Yang B, Qu J, Cheng Y, Zhu Z, Gao Y, Wang Q, Liu Y, Li Z, Qu X. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res. 2020;10:4513-4526. [PubMed] [Cited in This Article: ] |
| 198. | Jiang HJ, Huang YA, You ZH. SAEROF: an ensemble approach for large-scale drug-disease association prediction by incorporating rotation forest and sparse autoencoder deep neural network. Sci Rep. 2020;10:4972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 199. | Que SJ, Chen QY, Qing-Zhong, Liu ZY, Wang JB, Lin JX, Lu J, Cao LL, Lin M, Tu RH, Huang ZN, Lin JL, Zheng HL, Li P, Zheng CH, Huang CM, Xie JW. Application of preoperative artificial neural network based on blood biomarkers and clinicopathological parameters for predicting long-term survival of patients with gastric cancer. World J Gastroenterol. 2019;25:6451-6464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 200. | Li C, Yu H, Sun Y, Zeng X, Zhang W. Identification of the hub genes in gastric cancer through weighted gene co-expression network analysis. PeerJ. 2021;9:e10682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 201. | Anekboon K, Lursinsap C, Phimoltares S, Fucharoen S, Tongsima S. Extracting predictive SNPs in Crohn's disease using a vacillating genetic algorithm and a neural classifier in case-control association studies. Comput Biol Med. 2014;44:57-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 202. | Fioravanti D, Giarratano Y, Maggio V, Agostinelli C, Chierici M, Jurman G, Furlanello C. Phylogenetic convolutional neural networks in metagenomics. BMC Bioinformatics. 2018;19:49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 203. | Hardalaç F, Başaranoğlu M, Yüksel M, Kutbay U, Kaplan M, Özderin Özin Y, Kılıç ZM, Demirbağ AE, Coşkun O, Aksoy A, Gangarapu V, Örmeci N, Kayaçetin E. The rate of mucosal healing by azathioprine therapy and prediction by artificial systems. Turk J Gastroenterol. 2015;26:315-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 204. | Klein A, Mazor Y, Karban A, Ben-Itzhak O, Chowers Y, Sabo E. Early histological findings may predict the clinical phenotype in Crohn's colitis. United European Gastroenterol J. 2017;5:694-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 205. | Peng JC, Ran ZH, Shen J. Seasonal variation in onset and relapse of IBD and a model to predict the frequency of onset, relapse, and severity of IBD based on artificial neural network. Int J Colorectal Dis. 2015;30:1267-1273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |